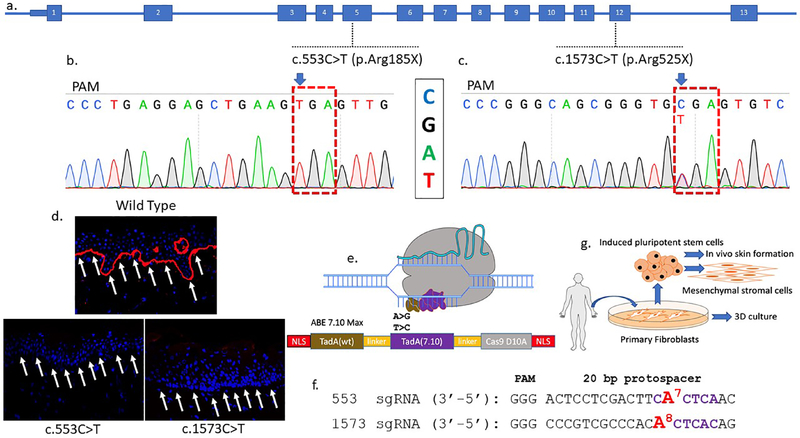
Figure 1. COL7A1 base editing experimental design.
a COL7A1 exons are numbered and the mutations targeted in this study are shown. One patient harbors a homozygous C:G>T:A mutation at nucleotide position c.553 causing an arginine to stop codon mutation in exon 5 at amino acid residue 185. The second patient was diagnosed as a compound heterozygote with a c.1573 C>T mutation in exon 12 at amino acid position 525 leading to a premature termination codon. The second allele has a c.2005C>T (R669X) mutation. b and c Sanger sequencing chromatograms of primary fibroblast samples from homozygous RDEB patients with Arg>X mutations. The sequence shown in the chromatograms corresponds to the sgRNA sequence and the NGG protospacer adjacent motif (PAM) sequence is indicated (anti-sense). d Skin biopsy immunofluorescence. Full thickness punch skin biopsies were obtained from a healthy control and the c.553 C>T and c.1573 C>T RDEB patients. Each sectioned tissue was stained with an equivalent amount of a polyclonal anti-C7 antibody and analyzed by confocal microscopy. C7 stains in red and white arrows show the DEJ in wild type but not RDEB samples. e Adenine base editor architecture. N- and C-terminal nuclear localization signals flank the E. coli TadA wildtype (WT) and evolved adenine deaminases that are separated by an XTEN linker and fused to the S. pyogenes Cas9 D10A nickase. Following Cas9 binding, the deaminases can act on the single-stranded DNA displaced by the protospacer. f c.553 and c.1573 sgRNA sequences with the target base numbered and shown in red. Purple lettering shows the ABE activity window between nucleotides corresponding to positions 4–8 of the protospacer (counting the 5’ nucleotide as position 1). g RDEB primary fibroblasts and induced pluripotent stem cells (iPSC) were obtained and corrected by electroporation of base editor mRNA and targeting sgRNAs. Fibroblasts were used in 3D organotypic cultures in vitro. Corrected iPSC were used as a platform for mesenchymal stromal cell derivation and in vivo teratoma formation that gave rise to ectoderm derived skin.