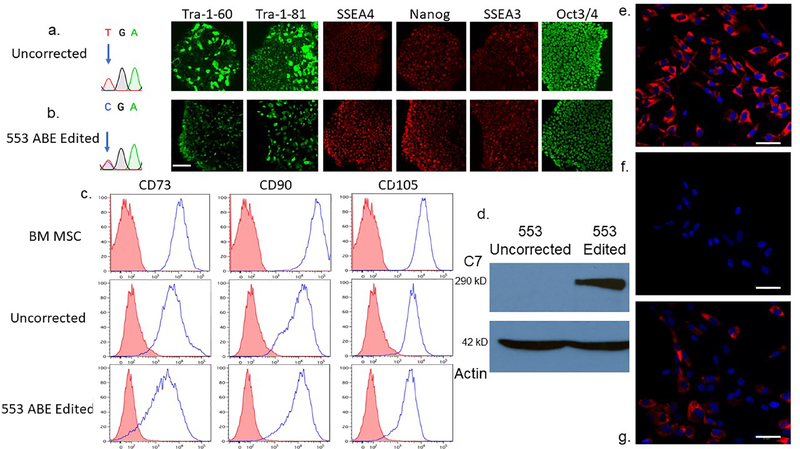
Figure 5. RDEB induced pluripotent stem cell base editing and directed mesenchymal stromal cell differentiation and characterization.
a and b iPSC editing. Sanger chromatogram of uncorrected c.553 C>T; R553X iPSC and a representative base edited clone are shown with arrow showing the mutant/target base. B Pluripotency immunofluorescence marker analysis. Antibodies against the pluripotency markers: podocalyxin TRA-1–60/TRA-1–81, Stage-specific embryonic antigen 4 (SSEA4), SSEA3, Nanog and OCT3/4 were used to detect expression levels. c Mesenchymal stromal cell characterization. Adult bone marrow from normal donors or iPSC derived MSCs were analyzed for the cell surface markers CD73, CD90, and CD105. The isotype staining control peaks are shown in pink. d iPSC derivative MSC Western blot analysis. MSCs derived from uncorrected c.553 C>T iPSCs were compared with those corrected by ABE in a Western blot using a polyclonal anti-C7 antibody. A ~290 kD band is shown with a 42 kD actin loading control below. e-g C7 immunostaining of chamber slides containing e wild type, bone marrow derived MSCs, f unedited 553 patient RDEB iPSC-derived MSCs, and g ABE edited 553 iPSC derivative MSCs. All cells were stained at the same time with the equivalent amount of polyclonal anti-C7 primary and secondary antibodies. Scale bar=50 μm.