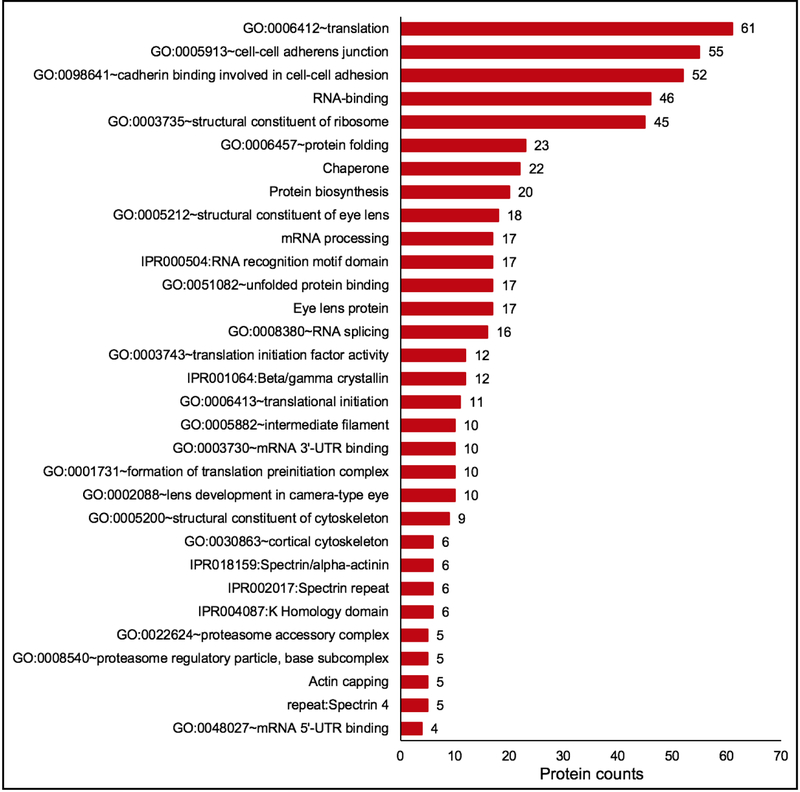
Fig. 6. Gene ontology (GO) analysis of lens enriched proteins identifies candidates relevant to function in the lens.
Lens enriched proteins were subjected to cluster-based analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID v6 .8) for functional annotation by gene ontology (GO) categories. This analysis assigned 406 lens enriched proteins into 68 annotation clusters. The top clusters included GO categories that are relevant to lens biology and cataract. These were “eye lens protein”, “protein folding”, “Ribonucleoprotein”, “Protein biosynthesis”, and “cell-cell adherens junction”, among others. Within the cluster for “eye lens protein”, other lens-relevant sub-categories were identified such as “structural constituent of eye lens”, “Beta/Gamma crystallin”, “lens development in camera-type eye”, “eye development” and “visual perception”. The number on the top of the bar graph shows the number of identified proteins in the category.