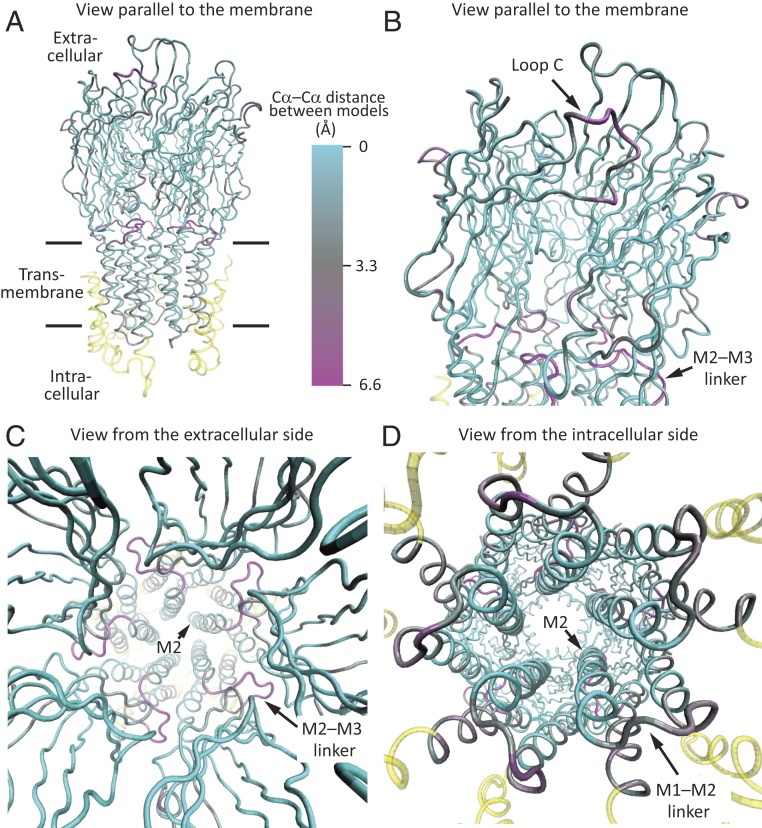
Fig. 2.
Cryo-EM structures of unliganded and propylammonium-bound ELIC embedded in POPC-only nanodiscs. The two models were superposed “globally” in such a way as to minimize the root mean-square Cα–Cα distance between identically numbered residues using the stretch ranging from Pro-11 (the first amino acid in the structure) to Ile-280 (at the C terminus of M3) of all five subunits. Shown is only the propylammonium-bound model, displayed in ribbon representation, and colored according to Cα deviations from the model of POPC-embedded unliganded ELIC. (A and B) Views parallel to the plane of the membrane. The enlarged view in B emphasizes the extracellular domain and the ECD–TMD interface. (C) View perpendicular to the plane of the membrane, from the extracellular side. (D) View perpendicular to the plane of the membrane, from the intracellular side. The cryo-EM density for the M1–M2 linkers was weak. The color code in A is the same for all panels. Regions not included in the superposition are colored yellow. Molecular images were prepared with VMD (37).