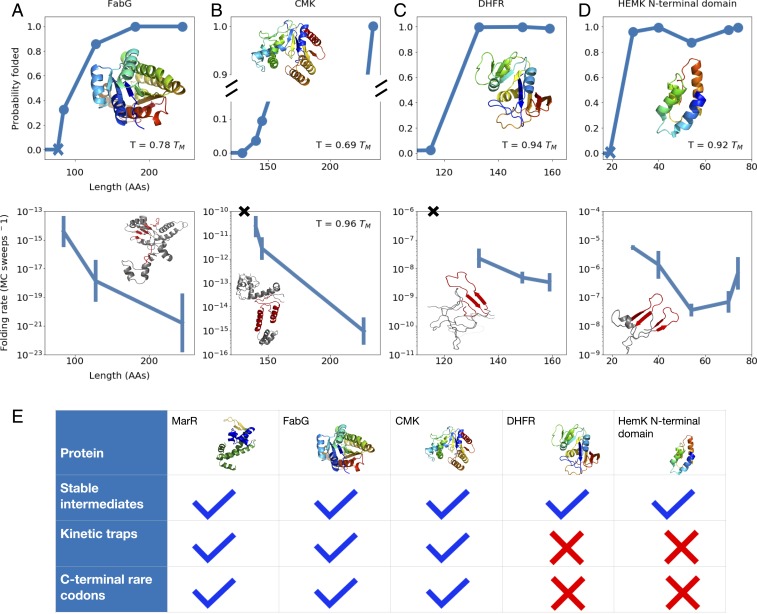
Fig. 5.
(A–D) As a function of chain length, the equilibrium probability that tertiary structure elements associated with the rate-limiting step are formed (Top) and the folding rate associated with the rate-limiting step (Bottom) are shown for proteins (A) FabG, (B) CMK, (C) DHFR, and (D) HemK. For each protein, the native structure (A–D, Top) and a sample structure that has yet to undergo the rate-limiting folding step (A–D, Bottom) are shown, with C-terminal nonnative contacts that must be broken prior to this step highlighted in red. Blue Xs in A and D, Top indicate the lengths at which the first amino acids associated with the rate-limiting step have been synthesized, while black Xs in B and C, Bottom indicate that no folding rate is computed because, even though enough residues have been synthesized for the rate-limiting structures to fold, their stability is low. As before, for each protein, we work at a temperature at which the fully synthesized chain shows a folding stability of 5 to 15 . For more details pertaining to each protein, see SI Appendix. (E) For each protein simulated, we indicate whether stable cotranslational folding intermediates are formed, deep kinetic traps slow folding, and conserved C-terminal rare codons are found in the sequence.