Fig. 1.
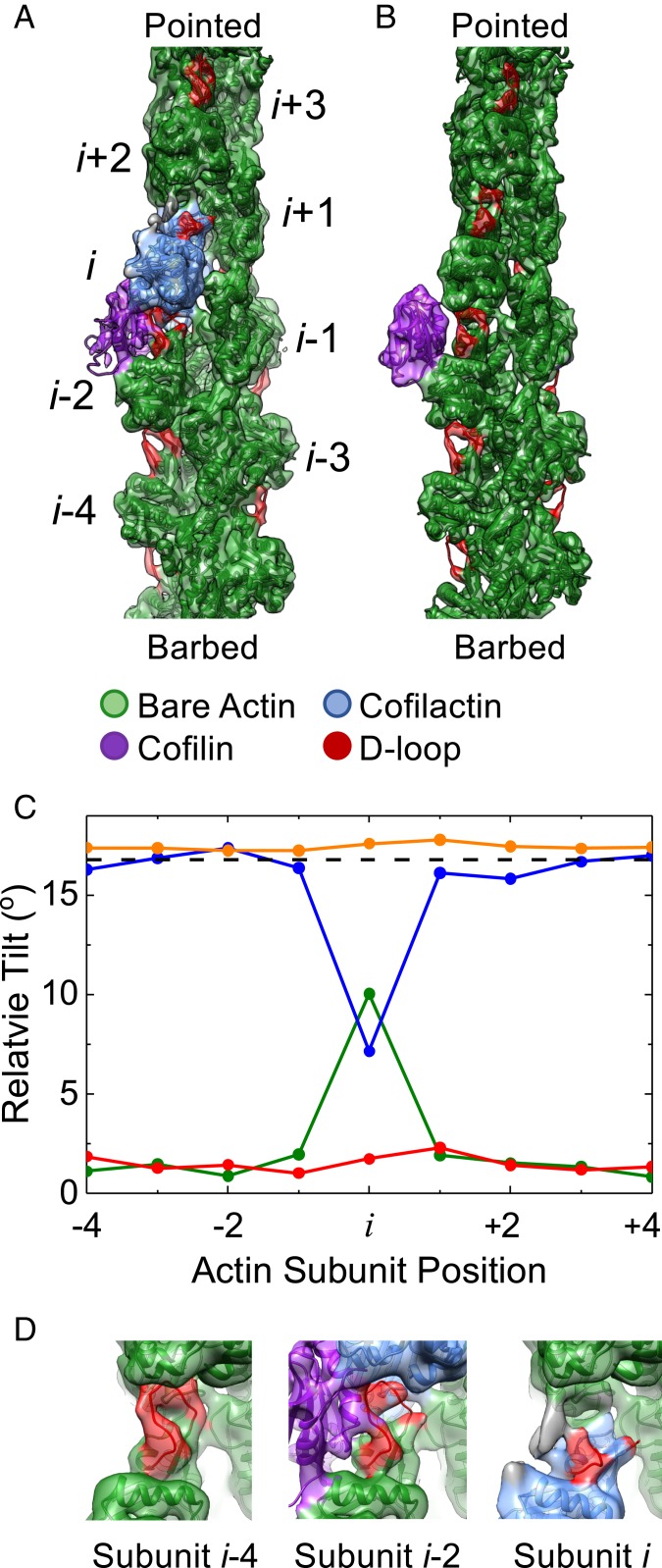
A single, bound WT cofilin disrupts intersubunit contacts between the actin subunit to which it is bound (subunit i) and its nearest longitudinal neighbor (subunit i − 2). (A) Cryo-EM density map reconstructed from filament segments (n = 8,917) containing ≥5 bare sites, followed by 1 cofilin-occupied site, followed by ≥4 bare sites progressing from the barbed to the pointed end of the filament. PDB models of actin and cofilactin were fit into the cryo-EM density. The cofilactin models are colored blue (PDB ID code 5YU8), bare actin is colored green (PDB ID code 6DJO), and cofilin is colored purple (PDB ID code 5YU8). The actin D-loop is colored red. (B) Cryo-EM reconstruction of 24,000 isolated, bound S3D-cofilin molecules identified by 3D classification on 5 actin subunit-long segments, colored as in A. (C) Fitted outer domains into the WT and S3D-cofilin maps were compared with reference cofilactin (WT, blue; S3D, orange) or bare actin (WT, green; S3D, red) models that were aligned to the actin inner domain region of the map. The rotation angle between the outer domains of the reference models is also shown (dashed black line). Only the actin subunit bound to WT cofilin toward the pointed end of the filament (subunit i) adopts a cofilactin-like conformation; all other subunits resemble bare actin. (D) Close-up view of the D-loops from actin subunits i, i − 2, and i − 4 from the single, bound WT cofilin fragment from identical orientations. Cofilin disrupts the D-loop cryo-EM density of the 2 actin subunits adjacent to it (subunits i and i − 2) while the D-loop cryo-EM density of all other D-loops remains properly positioned and relatively strong. Some unaccounted additional density (gray) appears near the D-loop of subunit i, which may originate from conformational mixing with an alternative structural state where cofilin is incompletely bound and/or fails to tilt subunit i.