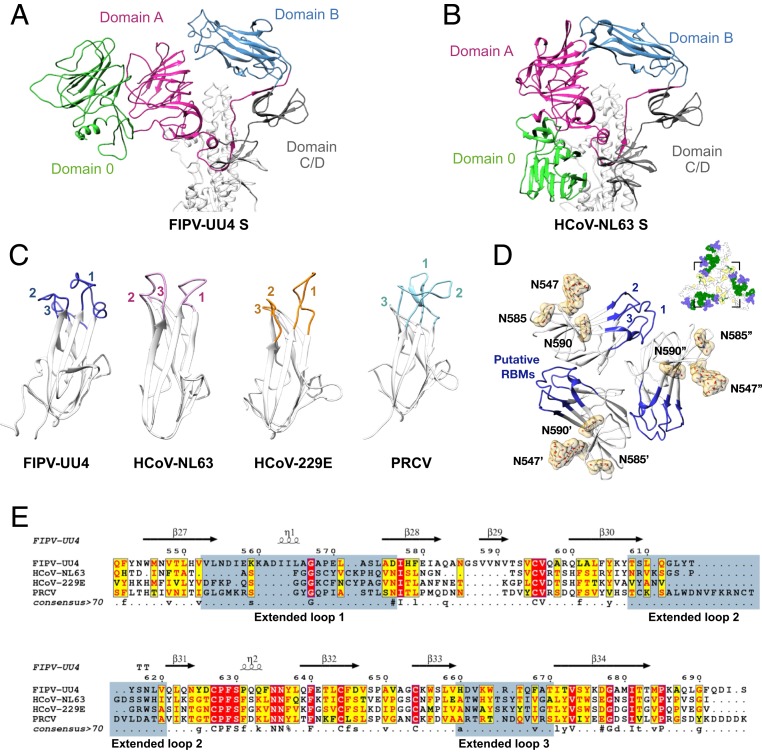
Fig. 4.
Structure divergence of host receptor recognition motifs among different CoVs. (A and B) Cartoon representations of FIPV-UU4 (A) and HCoV-NL63 (B) S1 subunits. The individual domains are colored in accordance with Fig. 1B. Note that domain 0 exhibits major conformational rearrangements between FIPV-UU4 and HCoV-NL63. (C) Structural comparison of domain B of FIPV-UU4, HCoV-NL63 (PDB ID code 3KBH), HCoV-229E (PDB ID code 6ATK), and PRCV (PDB ID code 4F5C). Extended loops 1 to 3 that are putative host receptor recognition motifs are highlighted with different colors and indicated with the respective numbers. (D) Expanded top view of the putative receptor-binding motifs of FIPV-UU4. The only 3 N-glycans in domain B are located on N547, N585, and N590, all of which are distant from the putative RBMs. A schematic view of the overall structure (the same as in Fig. 2B) indicates the absence of N-glycans in the putative RBMs. All N-glycan densities were segmented from the 3.3-Å map. (E) Sequence alignment of the domain B regions of FIPV-UU4 (residues 541 to 695), HCoV-NL63 (residues 481 to 616), HCoV-229E (residues 290 to 435), and PRCV (residues 283 to 430). Regions that correspond to the 3 extended loops are highlighted in blue.