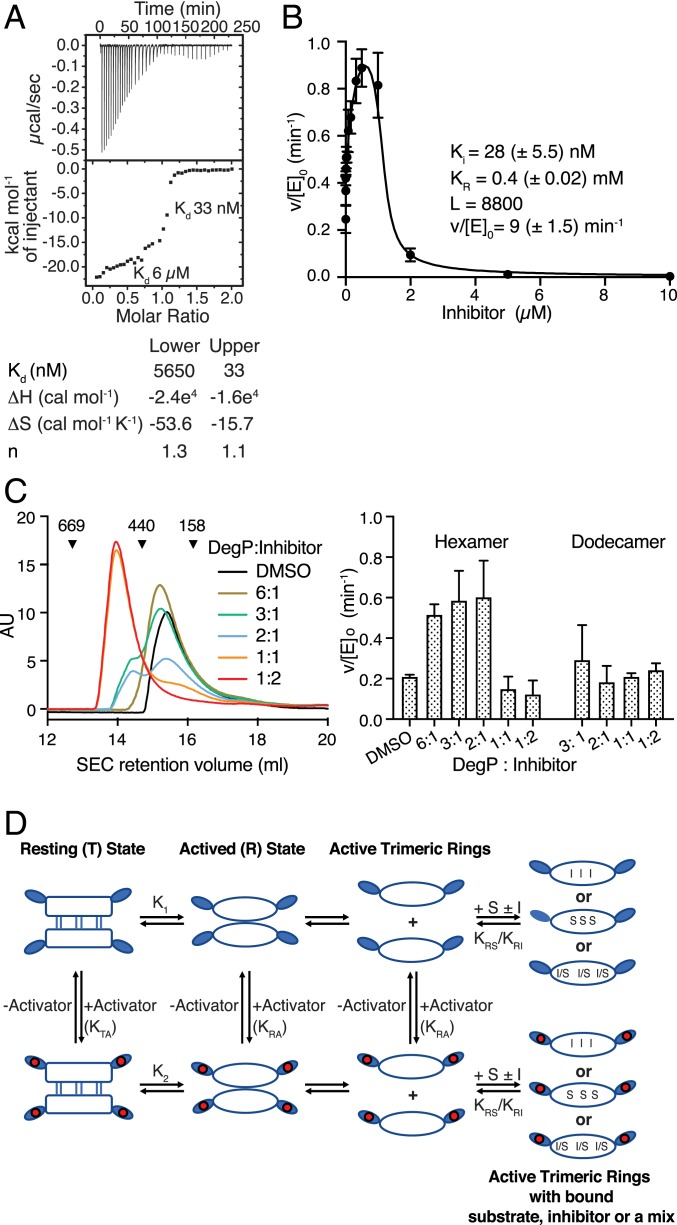
Fig. 1.
Inhibition of DegP by DPMFKLV-B(OH)2. (A) ITC thermogram of binding of DPMFKLV-B(OH)2 inhibitor to wild-type (wt) DegP. Here, 400 µM inhibitor was titrated into the sample cell containing 40 μM DegP. Kd of the two distinct binding steps are indicated. (B) Kinetic parameters of DegP (1 µM) activity in the presence of the DPMFKLV-B(OH)2 concentration indicated using the chromogenic substrate SPMFKGV-pNA (500 µM). Error bars = SD of experimental data (n = 3). Data were fitted to the Monod-Wyman-Changeux (MWC) model as described in SI Appendix. L, equilibrium constant T/R in the absence of ligand; Ki, affinity of the inhibitor to activated DegP; KR, dissociation constant for substrate binding to activated DegP; v/[E]0, maximum turnover rate for the activated DegP species; parentheses, 95% confidence limit of the fit. (C) size-exclusion chromatography (SEC) analysis of DegP (100 µM) preincubated with the ratios of DPMFKLV-B(OH)2 indicated. Retention volumes and size (kDa) of calibration proteins thyroglobulin (669 kDa), ferritin (440 kDa), and aldolase (158 kDa) are indicated (Left). Activity of eluted DegP fractions was determined using 500 µM SPMFKGV-pNA as substrate (Right). Error bars indicate SD (n = 3). (D) Cartoon of the core allosteric mechanism. The T state is the inactive hexamer. It exists in equilibrium with the dissociated trimers (R state), which is competent to bind either substrate (S) or inhibitor (I) to the active site. Although the equilibrium in the absence of S or I lies toward the T state (i.e., K1 is small), the binding of S or I pulls the equilibrium toward the R state by thermodynamic coupling. Alternatively, the inactive T state binds activator to the PDZ1 domains moderately, but the R state PDZ1 domains bind activator more tightly (i.e., KRA >> KTA), leading to activation because once in the R state it can bind S or I.