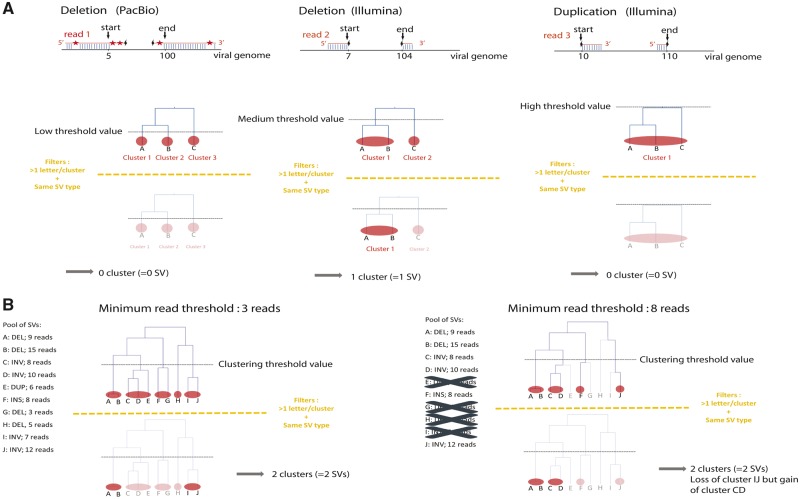
Figure 1.
Illustration of two important steps of the hierarchical clustering of SVs. (A) Influence of the clustering threshold value. The top panel illustrates three reads (one long PacBio read and two short Illumina reads) mapped onto overlapping regions of the viral genome. Red asterisks correspond to sequencing errors that prevent accurate mapping of long reads. ‘start’ and ‘end’ correspond to start and end coordinates of the SV detected by SV callers (a deletion in the case of Reads 1 and 2 and a duplication for Read 3. The bottom panel shows how using multiple clustering thresholds prevents discarding well-supported SVs. With a low threshold, all clusters contain a single SV because none of the SVs have the exact same coordinates. Because a downstream filter of our pipeline requires that SVs must be detected either by both long and short reads (in the case of the AcMNPV population sequenced using both Illumina and PacBio technologies) or by two programs (in the case of the three other large dsDNA viruses sequenced only with Illumina) to be retained, none of the SVs are retained with this low clustering threshold. With a high clustering threshold, all SVs (two deletions and one duplication) end up in the same cluster because they are defined by coordinates that are close to each other. Because a downstream filter of our pipeline requires that all SVs within a cluster must be of the same nature for a cluster to be retained, the cluster is here not considered further. With a medium threshold, the deletions detected by Reads 1 and 2 are lumped into the same cluster because their coordinates are close enough and the duplication detected by Read 3 forms another cluster because its coordinates are too far from those of the deletion. After running the downstream filters of our pipeline, Cluster 2 is retained and one deletion is counted because it has been detected independently by long and short reads. The cluster containing the duplication is not considered further because it contains only one SV detected by short reads only. Note that although SVs supported by only one read are represented here for the sake of simplicity but our approach only retained SVs supported by a minimum of three reads. (B) Influence of the minimum number of reads supporting a SV. On the left panel, using three reads as the minimum number of reads required to retain SVs, ten SVs of different nature and/or supported by different numbers of reads have been detected by SV callers. Under a given clustering threshold value, these ten SVs form five clusters, only two of which are retained (A–B and I–J) by downstream filters because they contain several SVs which are all of the same nature. On the right panel, only six of the ten SVs detected on the left panel are detected by SV callers using eight reads as the minimum number of reads required to retain SVs. With the same given clustering threshold value as in the left panel, SVs form four clusters, two of which (A, B and C, D) are retained by downstream filters because they contain several SVs which are all of the same nature. Using multiple minimum numbers of reads supporting SVs ensure that well-supported SVs (here the inversion in C and D) are not eliminated by downstream filters.