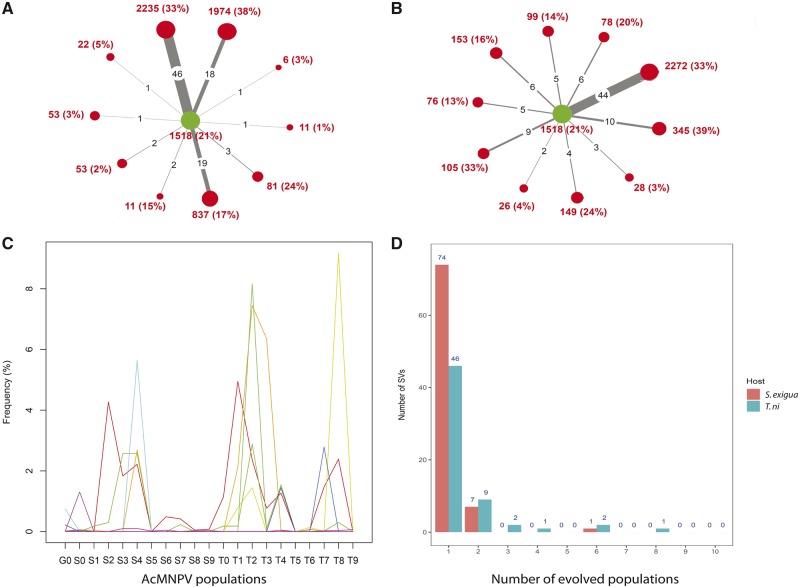
Figure 5.
Dynamics of SVs in twenty evolved AcMNPV lines. (A) Red circles show the number of SVs detected in ten AcMNPV populations which were each purified after ten infection cycles on larvae of the beet armyworm (S. exigua). The green circle shows the number of SVs detected in the parental population of AcMNPV purified from larvae of the cabbage looper moth (T. ni). The size of the circle is proportional to the number of SVs and the frequency of viral genomes carrying a SV is given between brackets, assuming the number of SVs per viral genome follows a Poisson distribution. The thickness of the lines linking the parental AcMNPV population to each of the ten evolved populations is proportional to the number of shared SVs (numbers in black on the lines). (B) Same as in A except that the ten evolved AcMNPV populations were purified after ten infection cycles on larvae of the same species (T. ni) as that used to generate the parental AcMNPV population. (C) Frequency of the ten most frequent SVs detected among the twenty-one viral populations and which were initially present in the parental AcMNPV population. One color corresponds to one SV. The ‘G0’ population refers to the parental population. The S0–S9 populations refer to the populations evolved on S. exigua larvae. The T0–T9 populations refer to populations evolved on T. ni. Note that no SVs reached >10 per cent in frequency in any AcMNPV population. (D) Number of SVs present in the parental AcMNPV population that were also detected in one to ten evolved viral populations. Most SVs were only detected in one evolved population (seventy-four in S. exigua and forty-six in T. ni).