Figure 5.
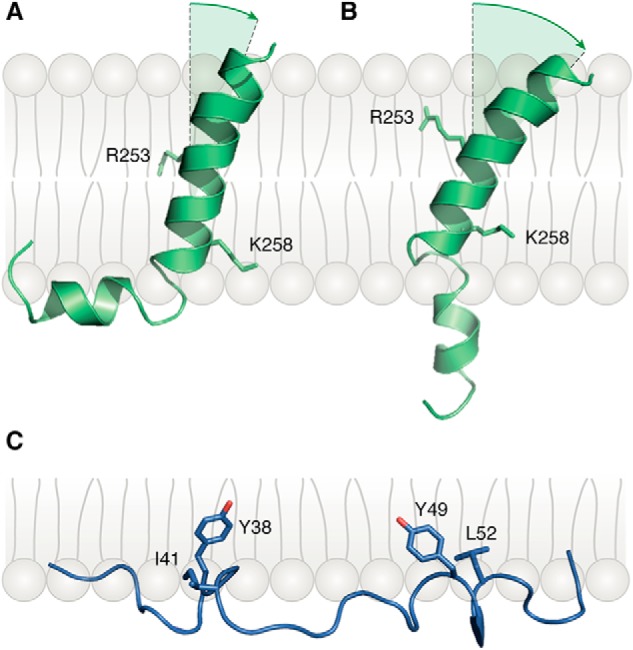
Bent and extended conformations of the TCRα TM helix. A, NMR structure of the TCRα TM helix in the bent conformation (PDB code 6MF8) (30). In the TCR–CD3 structure (23), TCRα Arg-253 and Lys-258 form salt bridges with acidic residues in CD3 TM helices (Fig. 2B). B, extended-state structure of the TCRα TM helix. C, conformation of the CD3ϵ cytoplasmic domain relative to the membrane (PDB code 2K4F) (29). The aromatic side chains of residues Tyr-38 and Tyr-49 of the CD3ϵ ITAM are embedded in the lipid bilayer.
