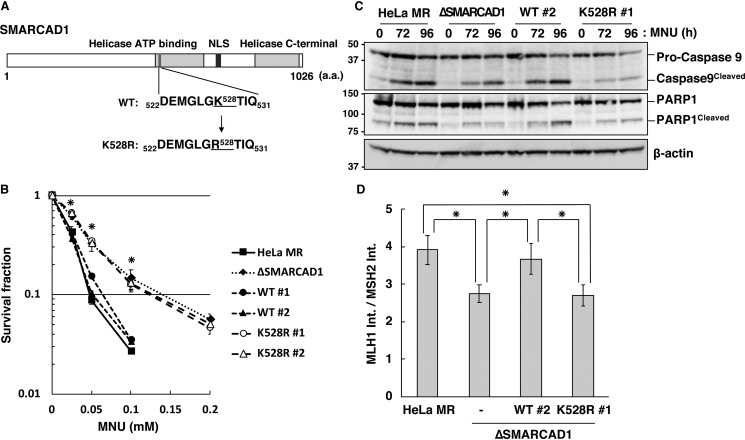
Figure 6.
Requirement of ATPase activity of SMARCAD1 for MMR-induced apoptosis. A, structure of human SMARCAD1 protein. The position of amino acid substitution K528R in the ATP-binding motif of the protein is indicated. B, survival fraction of cells expressing WT or mutant SMARCAD1 after MNU treatment. The cells were treated with MNU for 1 h; the number of colonies formed was then counted, and the survival fractions were determined. Values are the means of at least three independent experiments, and error bars indicate S.E. *, significant differences (p < 0.05). C, ATPase activity of SMARCAD1 relevant to apoptotic induction. Whole-cell extracts were prepared from cells treated with MNU and subjected to SDS-PAGE, followed by immunoblotting using anti-caspase-9 and anti-PARP1 antibodies. β-Actin was the loading control. The molecular weights (× 10−3) are indicated on the left of the panels. D, ATPase activity of SMARCAD1 relevant to the formation of MMR complex. The relative amounts of MLH1 co-immunoprecipitated with MSH2 after MNU treatment were calculated as described in the legend to Fig. 5D and are represented as means ± S.E. of six independent experiments. *, significant differences (p < 0.05).