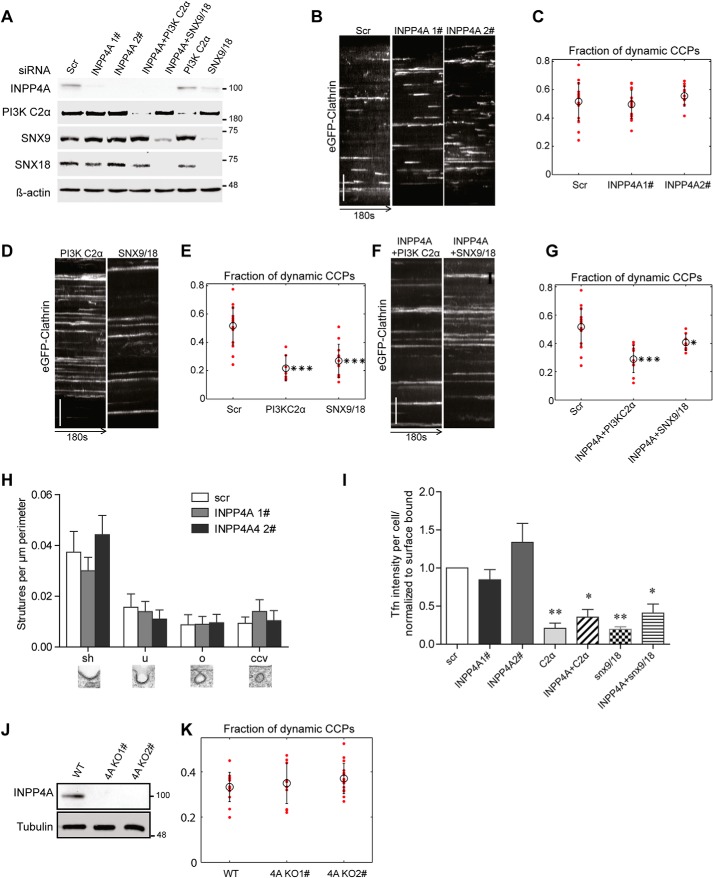
Figure 5.
INPP4A is dispensable for CME. A, immunoblot analysis of COS7 cells transfected with the indicated siRNAs confirms efficient depletion of their targets. β-Actin was analyzed as a loading control (INPP4A 2 was used for co-knockdown with snx9 and PI3KC2α). Scr, scrambled. B, representative kymographs of eGFP-clathrin puncta over 180 s in control cells (treated with scrambled siRNA) or cells depleted of INPP4A by two different siRNAs. y axis scale bar = 5 μm. C, fraction of dynamic CCPs in control cells (Scr) or cells depleted of INPP4A by two different siRNAs, analyzed by quantitative automated 2D tracking. D and F, kymographs of eGFP-clathrin puncta over 180 s in control cells (Scr) or cells depleted of PI3KC2α or SNX9/SNX18 either alone (D) or together with INPP4A (F). y axis scale bars = 5 μm. E and G, fraction of dynamic CCPs in control cells (Scr) or cells depleted of PI3KC2α or SNX9/SNX18 either alone (E) or together with INPP4A (G), analyzed by quantitative automated 2D tracking. H, ultrastructural analysis of endocytic intermediates in control cells (Scr) or cells depleted of INPP4A by two different siRNAs by morphometric quantification of thin-section electron microscopy images. The different morphological groups are indicated below the bar diagram. I, depletion of INPP4A by two different siRNAs does not impair CME of transferrin. Bar diagrams represent the ratio of internalized (10 min, 37 °C) to surface-bound transferrin (45 min, 4 °C). Mean ± S.E., n = 3 independent experiments. 1202, 633, 761, 559, 645, 764, and 464 cells from three independent experiments were analyzed. *, p < 0.05; **, p < 0.01; one-way ANOVA. J, loss of INPP4A expression in INPP4A KO COS7 cells. Tubulin was analyzed as a loading control. Numbers in A and J indicate the molecular mass of the immunoblotted proteins in kilodaltons. K, fraction of dynamic CCPs in control or INPP4A KO COS7 cells, analyzed by quantitative automated 2D tracking.