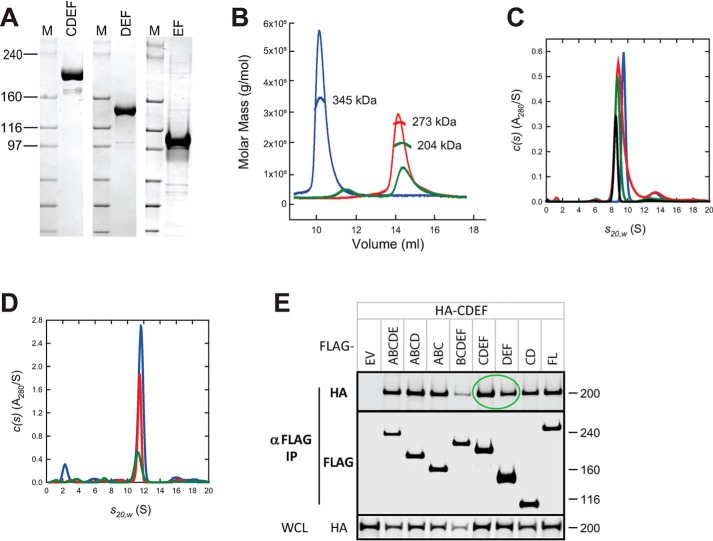
Figure 5.
C-terminal domains of neurofibromin are capable of dimerization. A, SDS-polyacrylamide gel of purified CDEF, DEF, and EF proteins. Size of molecular mass markers are noted in kilodaltons. B, SEC-MALS analysis of CDEF (blue), DEF (red), and EF (green) proteins. C, sedimentation velocity absorbance c(s) profiles for the DEF protein at 2.1 μm (green), 4.2 μm (red), and 8.8 μm (blue) based on data collected at 280 nm. D, sedimentation velocity absorbance c(s) profiles for the CDEF protein at 1.2 μm (black), 2.6 μm (green), 5.2 μm (red), and 10.5 μm (blue) based on data collected at 280 nm. Data at the highest concentration was collected using a 3-mm pathlength cell, and standard 12-mm cells were used for the lower concentrations. E, Western blotting of co-immunoprecipitation of differentially epitope-tagged NF1 proteins in HEK293 cells. In this figure, HA-CDEF protein was co-IPed with FLAG-tagged domains as noted. The top two gel sections are lysates purified with anti-FLAG antibodies and probed with antibodies to the HA or FLAG epitopes. The bottom section contains WCL probed with anti-HA antibodies. Molecular mass of standards is noted on the right in kilodaltons. Circled regions are discussed in more detail in the text. EV, empty vector control; FL, full-length NF1.