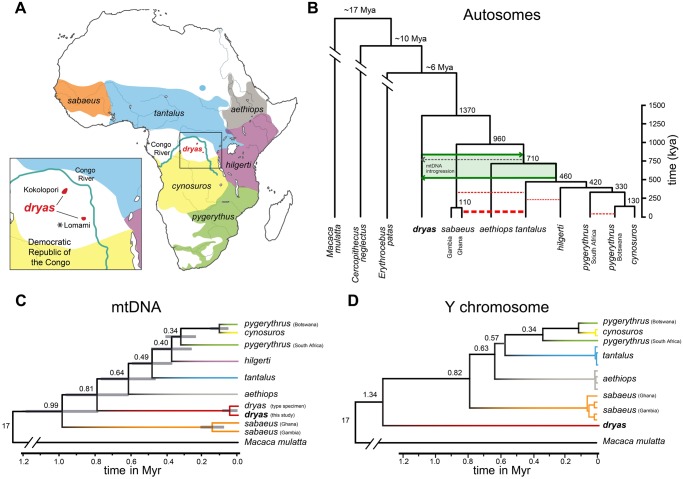
Fig. 1.
Phylogeny of the Dryas monkey and the vervets. (A) Distribution ranges of vervets. Cercopithecus dryas is currently known from only two isolated populations: The Kokolopori–Wamba and the upper basins of the Lomami and Lualaba rivers. The sequenced Dryas monkey individual was sampled from the Lomami population as indicated by the asterisk. (B) The autosomal consensus phylogeny and MCMC dating of the Dryas monkey and the vervets in comparison to two other guenon genomes, using the rhesus macaque (Macaca mulatta) as outgroup. The tree topology is supported by multiple analyses (see Materials and Methods and supplementary figs. S2 and S3, Supplementary Material online). Red dotted lines depict previously reported admixture events between the different vervet species (Svardal et al. 2017), with the line width corresponding to the relative strength of admixture. Green arrows show the best-supported timing of admixture between the Dryas monkey and the vervets. Chlorocebus sabaeus individuals from Ghana have a higher proportion of shared derived alleles with the Dryas monkey than Chl. sabaeus individuals from Gambia, which is the result of a strong secondary admixture pulse between Chl. tantalus and the Ghana Chl. sabaeus population (Svardal et al. 2017). Numbers at the nodes represent estimated mean divergence times (see supplementary fig. S3, Supplementary Material online). (C) Mitochondrial phylogeny and divergence times, provided as numbers at the nodes. Gray blocks show the 95% confidence intervals as obtained with MCMCTree. (D) UPGMA Y-chromosomal phylogeny, calibrated using the rhesus macaque Y-chromosome as outgroup. Number at the nodes represent estimated divergence times.