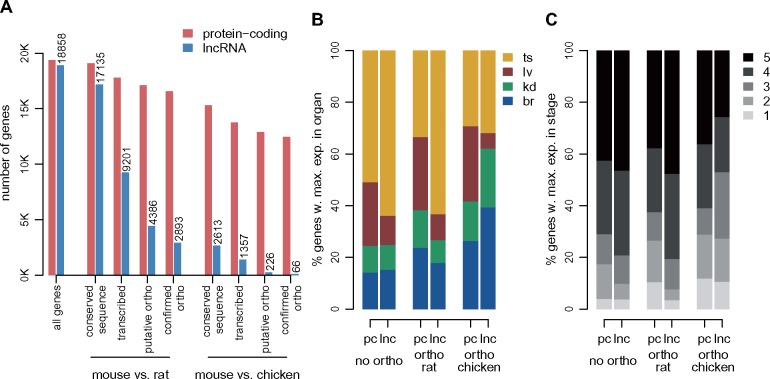
Fig. 5.
Orthologous lncRNA families for mouse, rat, and chicken. (A) Number of mouse protein-coding genes and lncRNAs in different classes of evolutionary conservation. From left to right: all loci, loci with conserved sequence in the rat, loci for which transcription could be detected (at least ten unique reads) in predicted orthologous locus in the rat, loci with predicted 1-to-1 orthologs, loci for which the predicted ortholog belonged to the same class (protein-coding or lncRNA) in the rat, loci with conserved sequence in the chicken, loci for which transcription could be detected (at least ten unique reads) in predicted orthologous locus in the chicken, loci with predicted 1-to-1 orthologs, loci for which the predicted ortholog belonged to the same class (protein-coding or lncRNA) in the chicken. We analyze 19,356 protein-coding genes and 18,858 candidate lncRNAs in the mouse. (B) Distribution of the organ in which maximum expression is observed, for mouse protein-coding and lncRNA genes that have no orthologs in the rat or chicken, for genes with orthologs in the rat and for genes with orthologs in chicken. The sample in which maximum expression is observed is computed based on an average expression values across biological replicates, for each organ/developmental stage combination (supplementary methods, Supplementary Material online). (C) Same as (B), for the distribution of the developmental stage in which maximum expression is observed.