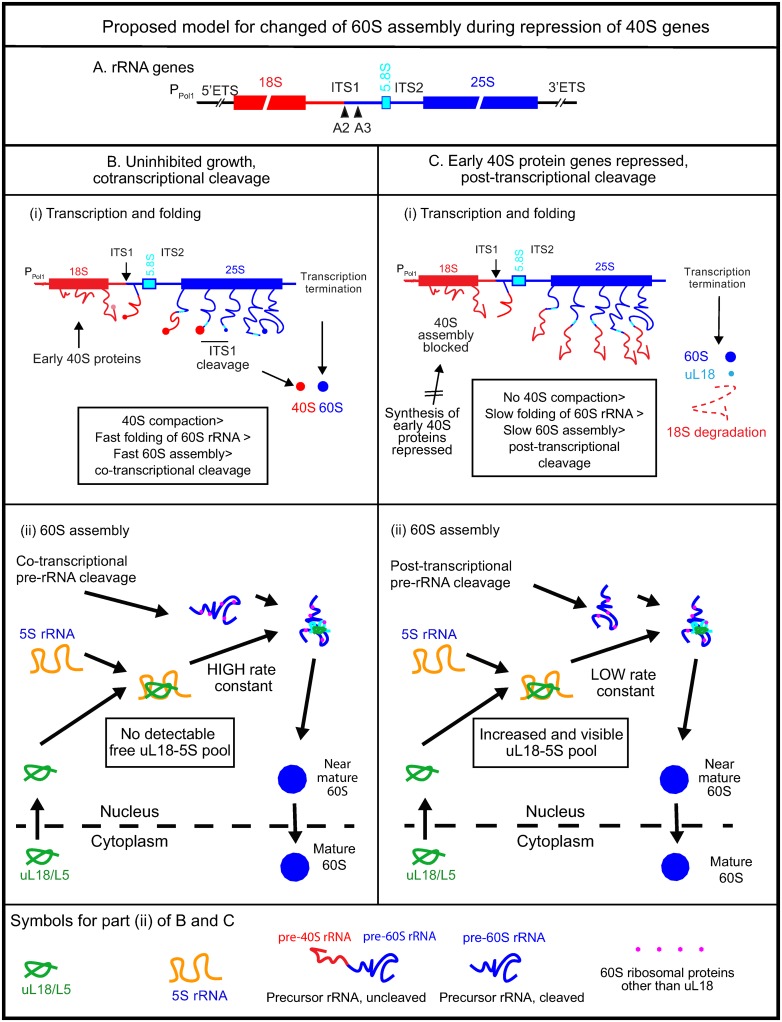
Fig 8. Model for effect of early 40S assembly on the kinetics of 60S assembly.
(A) Map of the long rRNA precursor gene transcribed by RNA polymerase I. ETS: External Transcribed Spacer; ITS: Internal Transcribed Spacer; A2 cleavage site separating the pre-rRNA into subunit-specific parts. (B) Co-transcriptional rRNA cleavage during unrestricted 40S assembly. (i) Schematic representation of co-transcriptional rRNA cleavage and ribosomal assembly as revealed by electron micrographs (Miller Spreads) [30]. The 18S parts of the transcript is in red, 5.8S in turquoise, and 25S in blue. “Wiggled” lines: uncompacted rRNA complexed with assembly factors and early assembly r-proteins. Filled circles: Compacted rRNA-protein complexes; the intensity of the red color indicates the density (compaction) of the complex. (ii) Kinetic model for formation of the uL18-5S rRNA complex and incorporation into the pre-60S during unrestrained 40S assembly. We propose that the rate constant for uL18/5S rRNA binding to the pre-60S is high in this condition. Therefore, uL18 is rapidly incorporated into the pre- 60 and uL18 does not accumulate outside the ribosome. (C) Post-transcriptional cleavage during inhibited 40S assembly. (i) Schematic illustration of electron micrographs of rRNA genes during repression of 40S r-protein genes. Pre-rRNA cleavage happens after transcription termination and compaction of the pre-40S and pre-60S is essentially absent. We suggest that absence of 40S compaction delays cleavage and changes the folding of the 60S-specific rRNA. (ii) Kinetic model for formation of the uL18-5S complex and incorporation into the pre-60S. We propose that the affinity (rate constant) of uL18-5S rRNA binding to the pre-60S is reduced due to the alternate folding and, possibly, alternate assembly pathway during post-transcriptional cleavage. Therefore, the rate of incorporation of uL18 into the pre-60S is slow leading to formation of an extra-ribosomal uL18 pool.