Figure 7.
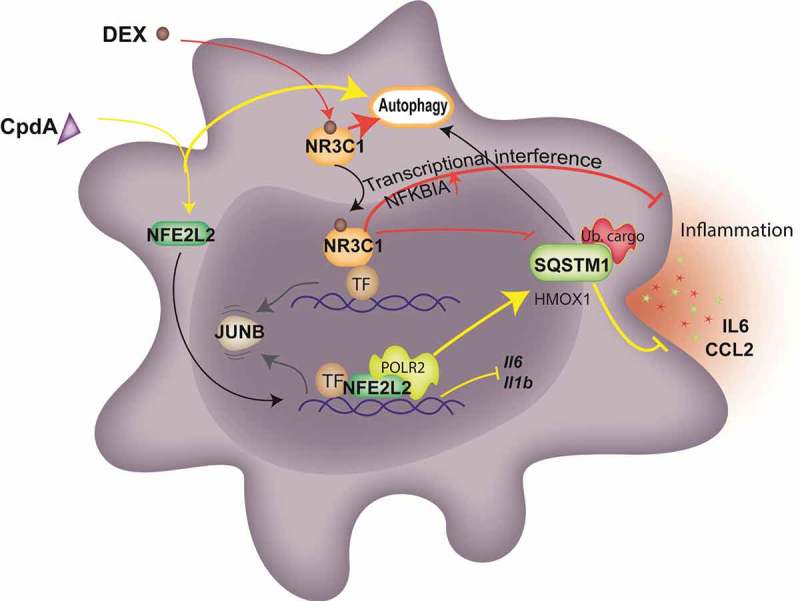
The model of NFE2L2- and NR3C1-dependent transcriptional regulation of SQSTM1 following CpdA and DEX and its link to autophagy and inflammation in macrophages. CpdA supports recruitment of NFE2L2 and a transcriptional, NR3C1-independent, upregulation of a subset of NFE2L2 pathway genes. One of those, encoding SQSTM1, is involved in CpdA-mediated suppression of inflammation. Oppositely, NFE2L2 recruitment at Il6 and Il1b promoters results in the downregulation of these genes. DEX mediates NR3C1-dependent transcriptional suppression of Il6 and Il1b genes as a main driver of its potent anti-inflammatory properties, yet is assisted by the upregulation of anti-inflammatory genes and the stabilization of NFKBIA/IκBα. The above-described transcriptional events result in a release of the transcription factor JUNB. Both compounds are able to induce autophagy with slightly different characteristics. CpdA leads to stronger ubiquitination of proteins at early time points, linking to aggregate-autophagy receptor binding [52]. Ub. cargo, ubiquitinated cargo; yellow lines depict activation or suppression by CpdA; red lines depict activation or suppression by DEX; black and gray arrows depict movement and sequence of events.
