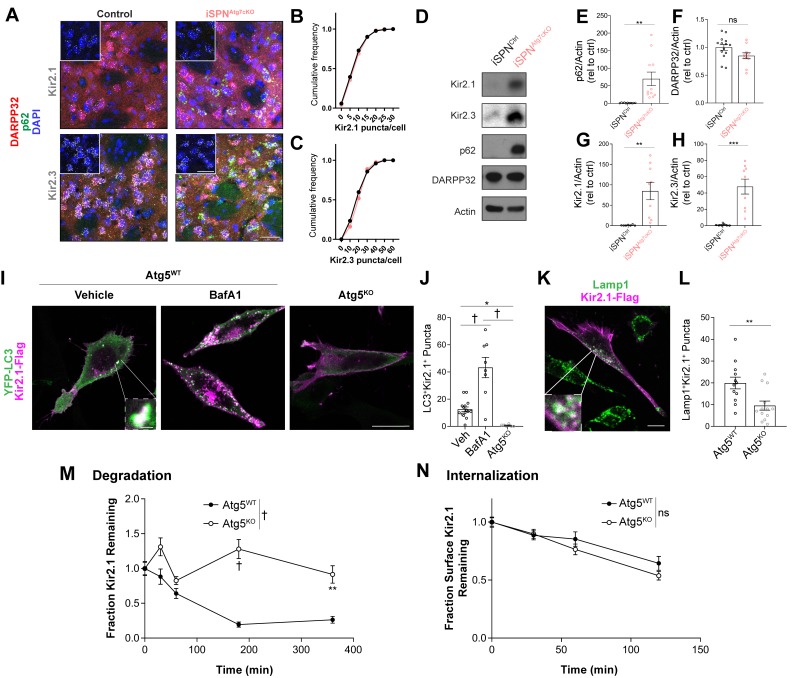
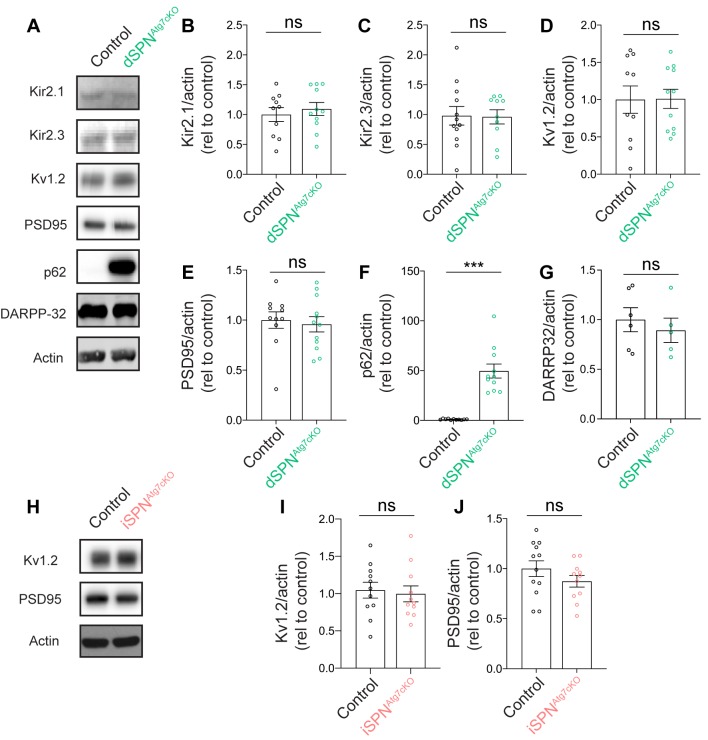
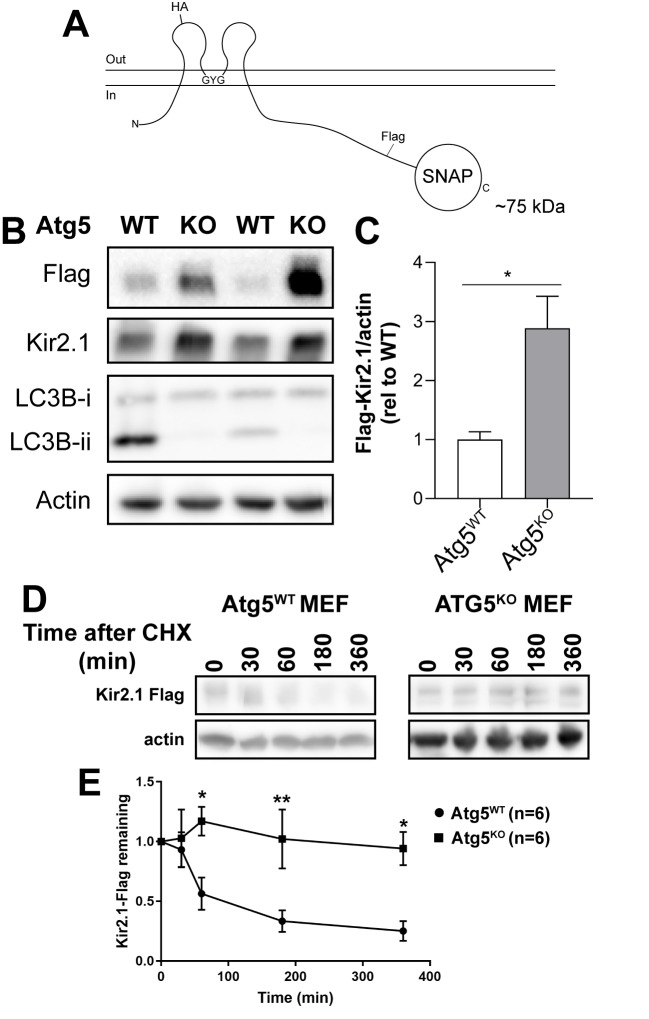
Figure 5. Autophagy is required for Kir2 degradation.
(A–C) There was no difference in Kir2.1 or Kir2.3 mRNA expression in SPNs of iSPNAtg7cKO mice in an RNAscope assay (N = 77–173 cells from three mice per group). Inset shows just RNAscope and DAPI. Scale bar 30 µm. Data analyzed by the Kolmogorov-Smirnov test, Genotype effect: p>0.05 for Kir2.1 and Kir2.3. (D) Representative western blots of specified proteins from total striatal lysates from iSPNCtrl or iSPNAtg7cKO mice. Quantifications of (E) p62 (t20 = 3.551, p = 0.0020), (F) DARPP32 (t23 = 1.984, p = 0.0593), (G) Kir2.1 (t14 = 3.435, p = 0.0040), and (H) Kir2.3 (t14 = 4.492, p = 0.0005) relative to actin. p62: iSPNctrl: n = 11, iSPNAtg7cKO: n = 11. DARPP32: iSPNctrl: n = 14, iSPNAtg7cKO: n = 11. Kir2.1: iSPNctrl: n = 7, iSPNAtg7cKO: n = 9. Kir2.3: iSPNctrl: n = 7, iSPNAtg7cKO: n = 9. Data analyzed by two-tailed, unpaired t test. (I-J) Kir2.1 is localized in LC3-GFP+ puncta in Atg5WT but not Atg5KO MEFs. BafilomycinA1 (BafA1; 100 nM 2 hr) treatment increases the number of LC3/Kir2.1-double labeled puncta in Atg5WT MEFs. Scale bar 20 µm. Inset scale bar 1 µm. Analyzed by one-way ANOVA followed by Bonferroni post-hoc test. F(2,26)=25.64, p<0.0001. (K-L) A reduction of Lamp1+Kir2.1+ puncta in Atg5KO MEFs. Scale bar 20 µm, inset scale bar 1 µm. Data analyzed by two-tailed, unpaired t test. t23 = 3.083, p = 0.0053. (M) Reduced degradation of SNAP-tag labeled Kir2.1 in Atg5KO MEFs. N: (WT,KO): T = 0 min (35,30), T = 30 min (30,27), T = 60 min (25,38), T = 180 min (33,67), T = 360 min (40,20). Data analyzed by two-way ANOVA. Genotype x time: F4,335 = 7.880, p<0.0001. (N) No significant difference in the internalization of antibody-labeled surface Kir2.1 channels in Atg5KO MEFs. N: (WT,KO): T = 0 min (76,56), T = 30 min (52,37), T = 60 min (41,42), T = 120 min (28,38). Data analyzed by two-way ANOVA. Genotype x time: F3,362 = 0.8038, ns; Time: F(3,362) = 25.88, p = 0.0001; Genotype: F(1,362) = 1.877, ns. ns p>0.05, *p<0.05, **p<0.01, ***p<0.001, † p<0.0001.