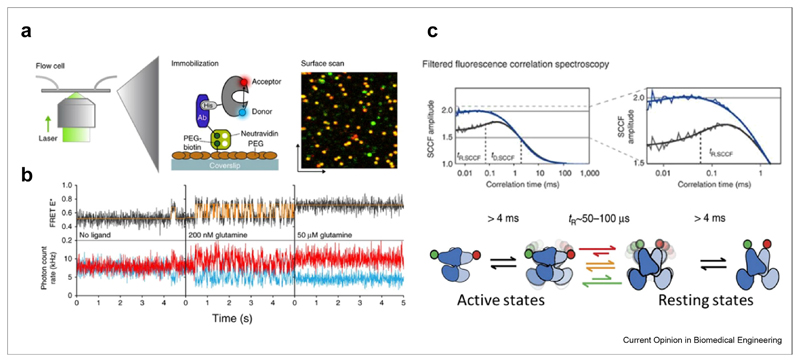
Figure 2. Membrane protein conformational dynamics measured with smFRET.
(a) Single-molecule dynamics of a surface-tethered SBD of the ABC importer GlnPQ probed by confocal scanning microscopy. (b) Single molecule trajectories at different ligand concentrations. Figure 2 A-B reprinted from Ref. [81] with permission from Nature publishing group. (c) Dynamics of the G-protein-coupled receptor mGluR, reprinted from Ref. [85] with permission. Top: filtered fluorescence cross-correlation curves, indicating microsecond conformational dynamics in the wild type (grey) but not in the constitutively active mutant (blue). Bottom: kinetic model of mGluR dynamics. SBD, substrate-binding domain; smFRET, single-molecule fluorescence resonance energy transfer; ABC, ATP-binding cassette.