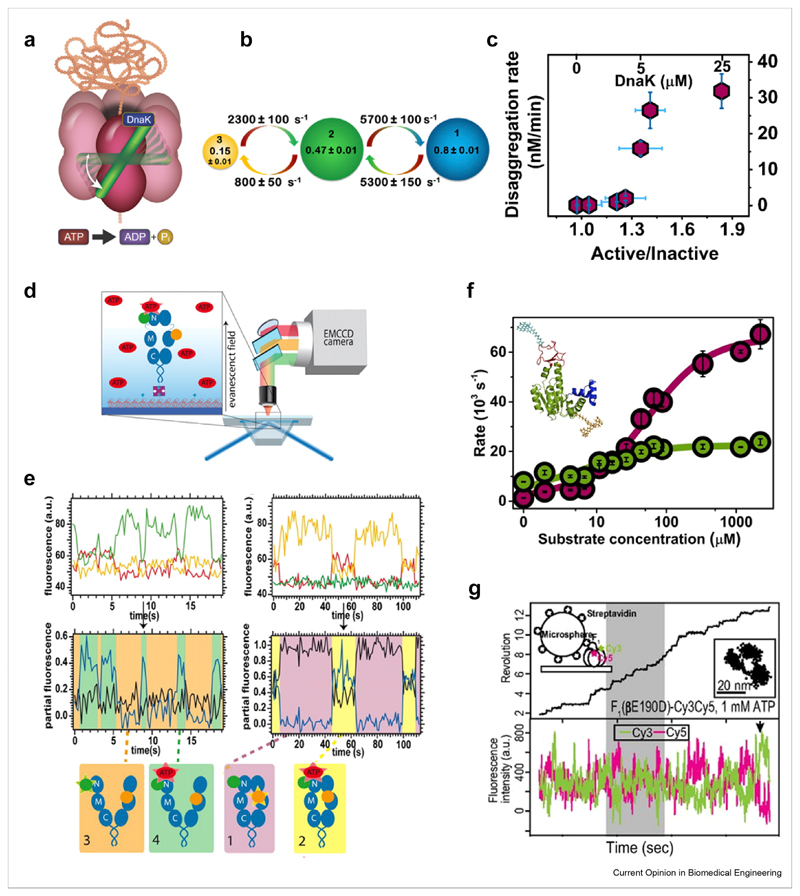
Figure 3. Dynamics of soluble protein machines measured with smFRET.
(a–c) The disaggregation machine ClpB, reprinted from Ref. [70] with permission. (a) Schematic of ClpB and its dynamic M domain (green), which toggles between three states on the microsecond timescale (b). (c) DnaK binding tunes the ratio between the two major states of the M domain and in turn disaggregation activity. (d–e) The chaperone Hsp90, reprinted from Ref. [47] with permission. (d) ATP-binding related dynamics of Hsp90 are studied using three-color smFRET. (e) Top: single molecule trajectories in three channels upon donor excitation. Middle: FRET efficiency trajectories in different states of Hsp90, as depicted in the bottom cartoons. (f) Adenylate kinase, reprinted from Ref. [33] with permission. Rates of domain closing and opening (cherry and green, respectively) as a function of substrate concentration. Inset: structure with attached labels. (g) F1-ATPase, Reprinted from Ref. [91] with permission. Simultaneous measurement of rotational motion of the ɣ-shaft using a bead (top panel, black line) and of FRET between αβ dimers (bottom panel, donor and acceptor in green and red). smFRET, single-molecule fluorescence resonance energy transfer.