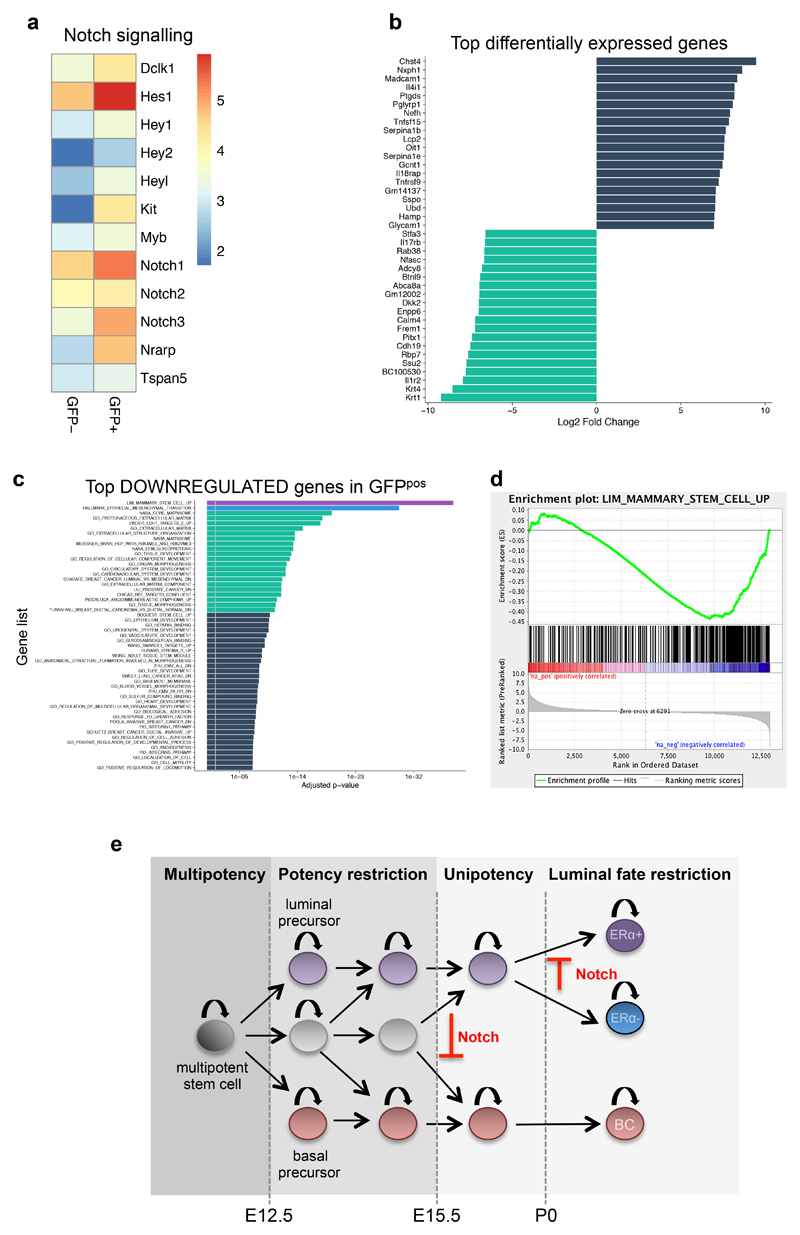
Figure 7. Transcriptomic analysis of the basal to luminal switch induced by Notch1 activation.
a. Heatmap of Notch1 related genes, using the log(fpkm) of the average expression values (fpkm) of 3 replicates each for GFPneg and GFPpos cells. b. Transcripts with more than five Log2 Fold Change (FC) differences obtained by RNA-seq of GFPneg and GFPpos sorted cells from SMA/N1ICD mice 72h after TAM induction; the blue bars correspond to the transcripts that are overexpressed, and the green bars indicate the transcripts that are downregulated. c. Over-represented GO categories among the top genes significantly downregulated upon Notch activation. d. Gene Set Enrichment analysis (GSEA) showing the inverse correlation between genes expressed upon Notch activation (in GFPpos cells) compared to the GO signature of “Mammary_Stem_Cell_Up”. A set of 12984 genes from three replicates was pre-ranked based on log2 fold changes. e. Model of the differentiation hierarchy during embryonic mammary gland development. Multipotent mammary stem cells are present in the early mammary placode, but at E12.5 some lineage restriction starts to occur, as unipotent luminal or basal precursor can be found at a frequency of about 30-40%, while the remaining MaSCs are still multipotent. Starting at embryonic day E15.5, no statistically discernable multipotency can be observed, suggesting that prenatal growth and branching of the mammary gland is supported by unipotent luminal or basal progenitors. At P0, an additional degree of fate restriction establishes two independently sustained luminal lineages, ERpos and ERneg luminal cells. Notch signalling prevents the generation of basal precursors during mammary embryogenesis and blocks ERpos cell fate specification.