Abstract
Donor-specific antibodies (DSAs) have a deleterious effect on allografts and remain a major immunologic barrier in transplantation. Current therapies to eliminate DSAs are ineffective in highly human leukocyte antigen (HLA)-sensitized patients. Proteasome inhibitors have been employed as a strategy to target bone marrow plasma cells (BMPCs), the source of long-term antibody production; however their efficacy has been limited by poorly defined drug resistance mechanisms. Here, we performed transcriptomic profiling of CD138+ BMPCs that survived in vivo desensitization therapy with the proteasome inhibitor carfilzomib to identify mechanisms of drug resistance. The results revealed a genomic signature that included increased expression of the immunoproteasome, a highly specialized proteasomal variant. Western blotting and functional studies demonstrated that catalytically active immunoproteasomes and the immunoproteasome activator PA28 were upregulated in carfilzomib-resistant BMPCs. Carfilzomib-resistant BMPCs displayed reduced sensitivity to the proteasome inhibitors carfilzomib, bortezomib and ixazomib, but enhanced sensitivity to an immunoproteasome-specific inhibitor ONX-0914. Finally, in vitro carfilzomib treatment of BMPCs from HLA-sensitized patients increased levels of the immunoproteasome β5i (PSMB8) catalytic subunit suggesting that carfilzomib therapy directly induces an adaptive immunoproteasome response. Taken together, our results indicate that carfilzomib induces structural changes in proteasomes and immunoproteasome formation.
1. INTRODUCTION
Proteasome inhibitors (PIs) have revolutionized treatment of the PC malignancy multiple myeloma (MM) changing MM from a uniformly fatal disease with limited life expectancy to a chronically manageable disease with substantially enhanced survival (1–3). Recently, PIs have been employed as a means to target non-transformed PCs that mediate autoimmune disease and HLA-sensitization in organ transplantation, where pathogenic antibodies (Abs) play a major role. Hence, PIs now provide the foundation as a therapeutic strategy to safely and effectively treat a number of PC-related human diseases (4).
Kidney transplantation is the treatment of choice for patients with end stage kidney disease (ESRD) as is associated with improved patient survival and better quality-of-life (5–7). It is now clear in kidney transplantation that Ab-mediated rejection (AMR) is the major cause of renal allograft loss and HLA Abs also promote AMR in heart, lung, and pancreas transplantation (8–10). HLA Abs, that arise from pregnancy, blood transfusion, or prior transplantation, present a significant and often impenetrable barrier to kidney transplantation, leading to increased morbidity and mortality (11, 12). Current desensitization therapies do not impact Ab production nor eliminate BMPCs (13–16). Transplantation across HLA barriers, through preconditioning with intravenous immune globulin (IVIg), alone or combined with plasmapheresis, or the CD20 monoclonal Ab rituximab provides short-term reductions in HLA Ab levels (17, 18). B-cell depletion with rituximab was shown to reduce preformed HLA Abs in only 10% of treated patients and failed to target BMPCs. Thus, allosensitization represents an urgent and unmet medical need in organ transplantation as current desensitization methods fail to provide durable effects on HLA Ab responses. Desensitization strategies that target BMPCs and lead to durable responses would allow highly sensitized patients to receive a donor organ.
BMPCs exhibit exceptionally high levels of immunoglobulin synthesis that is accompanied by the accumulation of endoplasmic reticulum stress. As a result, BMPCs are highly susceptible to agents that disrupt protein homeostasis, such as proteasome inhibition (19, 20). Bortezomib is a reversible PI that effectively targets malignant PCs and is used to treat newly diagnosed and relapsed/refractory MM. Carfilzomib is an irreversible, epoxyketone PI that functions similarly to bortezomib primarily through inhibition of proteasomal chymotrypsin-like (ChT-L) activity. Carfilzomib selectively inhibits the proteasome through a binding mechanism distinct from that of bortezomib, has a short half-life, and a lack of off-target effects which may explain the favorable safety profile compared to bortezomib.
Proteasomes are composed of four stacked heptameric rings in which non-identical, but structurally-related, α subunits form the two outer rings, and distinct β subunits form the two inner rings (21). Proteasomes can exist in a constitutive c20S form that exhibits peptide-hydrolyzing activities conferred by β1, β2 and β5 catalytic subunits (Supp. Fig. 1). c20S proteasome catalytic subunits can be substituted by the IFN-γ-inducible β1i, β2i and β5i catalytic subunits, encoded by PSMB9, PSMB10 and PSMB8, to form immunoproteasomes (i20S proteasomes) (22, 23). c20S proteasomes are capped at either or both ends by a 19S regulatory particle (RP) while i20S proteasomes are capped by a heptameric PA28α/β activator.
We and others have used PIs as a means to eliminate long-standing HLA Abs in transplant candidates, a process termed desensitization. Studies by our group and others have demonstrated that bortezomib consistently and substantially reduced HLA Ab levels in transplant candidates beyond that achieved with IVIg-based regimens (24–28). However, dose-limiting toxicity from peripheral neuropathy and the emergence of drug resistance has limited bortezomib efficacy in desensitization. Carfilzomib is clinically devoid of peripheral neuropathy but is also limited therapeutically by poorly defined chemoresistance mechanisms. Here, we hypothesized that carfilzomib treatment may induce proteasomal structural adaptations that contribute to drug resistance. We investigated the in vivo and in vitro effects of carfilzomib on BMPCs isolated from HLA-sensitized patients. Genomic analyses coupled with biochemical and functional assays, uncovered proteasomal adaptations that up regulate i20S proteasome levels and activity to reveal a novel mechanism of chemoresistance within the context of HLA-desensitization.
2. MATERIALS AND METHODS
2.1. BM aspiration and isolation of BMPCs.
BM was collected from the iliac crest using an aspiration needle under sterile conditions from patients enrolled in University of Cincinnati IRB-approved clinical trial NCT02442648. Patients were defined as HLA-sensitized if the calculated panel reactive Ab was >30% following single antigen bead (SAB, One Lambda, Canoga Park, CA) analysis on a Luminex® platform (Luminex, Austin, TX). Patient demographic features and clinical characteristics are provided in Supp. Table 1. Aspirates were diluted with sterile PBS, passed through a 100 uM filter to remove debris, layered onto 15 mL Ficoll-Paque plus (density 1.077 g/mL) and centrifuged at 445 x g. The pellet was removed, PBS added and centrifuged at 300 x g. To remove platelets, the pellet was resuspended in 50 mL PBS, centrifuged at 200 x g for 10 min and supernatant removed. The BM mononuclear fraction was then resuspended in PBS containing 0.5% BSA, 2 mM EDTA (buffer A), mixed with CD138 microbeads for 15 min at 4°C, and washed in buffer A. A CD138 column (Miltenyi Biotec, San Diego, CA) was equilibrated with buffer A at 4°C in the presence of a magnetic field, the suspension applied and the unbound CD138- fraction collected. The column was then removed from the magnetic field, placed in a sterile eppendorf tube and the CD138+ fraction collected (Supp. Fig. 2). Live CD138+ cells were isolated to >95% purity as indicated by trypan blue exclusion and flow cytometry.
2.2. Co-culture of BMPCs with BMSC media
BMPCs were isolated from BM biopsies performed prior to and after in vivo carfilzomib treatment of highly HLA-sensitized patients on protocol NCT02442648 (Supp. Fig. 3). CD138+ BMPCs were co-cultured with conditioned media from the human HS.5 BMSC line (ATCC, Manassas, VA) supplemented with APRIL and IL-6 for indicated times. HS.5 cells were grown to confluency and conditioned media was removed and centrifuged at 3,000 x g for 10 min.
2.3. Transcriptomic analysis of pre- and post-carfilzomib treated BMPCs
Total RNA was analyzed using an Ilumina® TruSeq® RNA Access Library kits (NGS, Illumina, San Diego, CA) platform (30 million reads/sample) with paired-end 75 bp sequencing to evaluate the effects of carfilzomib on gene expression. Strongly and differentially expressed gene sets were clustered and evaluated using enrichment and prior knowledge-based network analyses using ToppGene Suite (toppgene.cchmc.org/).
2.4. HLA Single antigen bead (SAB) assays
Patient serum, BM mononuclear cells, CD138+ and CD138- cells were incubated in 100 uL BMSC conditioned media at 37ΰC for 72 h. Serum or culture supernatants were then used for SAB assays. SAB assays were performed by incubating serum or culture supernatant with purified HLA antigen beads, washing, and a fluorescent-conjugated anti-human IgG Ab added, followed by analysis on the Luminex® platform. The platform generated a semi-quantitative output for each bead expressed as mean fluorescence intensity (MFI). The coefficient of variation for SAB assays was consistently between 5-8%.
2.5. Isolation of proteasomes from BMPCs
Proteasomes were isolated using agarose-immobilized 20S α7 (PSMA7) subunit-specific Ab (Enzo Life Sciences, Farmingdale, NY) that allows for rapid isolation of proteasome complexes from biological samples with high integrity and without genetic modification of the proteasome which is not feasible with patient-derived samples. Cells were washed with PBS, lysates prepared in non-denaturing buffer and incubated with Abs to PSMA7 at 4°C overnight with protein A sepharose. The slurry was washed with PBS and resuspended in non-denaturing loading buffer for native gels. The method maintains proteasome integrity, avoids steps that denature the proteasome as well as high salt buffers that dissociate proteasome-associated subunits.
2.6. SDS and native polyacrylamide gel electrophoresis
BMPCs isolated from healthy individuals, HLA-sensitized patients and HLA-sensitized patients treated with carfilzomib were lysed in buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β−glycerophosphate, 1 mM Na3VO4 and 1 <g/ml leupeptin). Lysed cells were incubated for 10 min at 4ΰC, centrifuged at 3,000 x rpm for 5 min and supernatant removed. SDS sample buffer was added, samples boiled for 5 min and loaded onto 4-12% bis-tris-polyacrylamide gels. Individual proteasome complexes isolated from patient BMPCs were resolved using 3-8% gradient native gel electrophoresis with tris-borate-EDTA buffer (TBE, 89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.3) as running buffer. Samples from healthy individuals, HLA-sensitized patients or HLA-sensitized patients that had received carfilzomib therapy were mixed with 4x loading buffer (100 mM Tris pH 8.0, 20% glycerol). Electrophoresis was carried out at 4°C in TBE running buffer with 0.5 mM DTT, 0.5 mM ATP and 2 mM MgCl2 added.
2.7. Western blotting
Proteins were transferred to PVDF membranes using a Bio-Rad mini-protean transfer system containing a buffer of 25 mM Tris base, 192 mM glycine, 0.1% SDS. Proteins were transferred for 6h at 40V at 4°C. Membranes with transferred proteins were blocked with a 1:1 mixture of Li-COR blocking buffer and PBS. Membranes were then incubated in blocking solution for 1 h and primary Abs (1:1000) applied for 16 h at 4°C. Membranes were washed and incubated with secondary Abs (IR Dye® 800CW goat anti-mouse IgG (H+L) or IR Dye 680CW donkey anti-rabbit IgG (H+L), and imaged using an Odyssey Li-Cor platform. Antibodies used were rabbit anti-PSMB5 polyclonal (Abcam, ab90867), rabbit monoclonal to anti-PSME1 (EPR10967, Abcam, ab155091), rabbit monoclonal to anti-PSME2 (EPR14931, Abcam, ab183727), rabbit anti-LMP2 polyclonal (PSMB9, Abcam, ab42987), rabbit anti-PSMB10 polyclonal (Abcam, ab77735), rabbit anti-LMP7 monoclonal (EPR14482, PSMB8, Abcam, ab180606), rabbit anti-PSMA6 polyclonal (Cell Signaling 2459), rabbit anti-PSMB1 polyclonal (Abcam, ab196623), rabbit anti-PSMB2 polyclonal (Abcam, ab140426), mouse anti-Rpn5 monoclonal (PSMD12, clone H3, Santa Cruz Biotechnology, sc-398279), mouse anti-α-tubulin monoclonal (DM1a, Abcam, ab7291), and rabbit anti-histone 3 polyclonal (Abcam, ab1791).
2.8. In situ proteasome activity overlay assay
Proteasomes (5ug/sample) were loaded onto native gels run as indicated above. Gels were incubated with either the pan-constitutive proteasome substrate Suc-LLVY-MCA (100uM) or the i20S proteasome substrate Ac-ANW-AMC (100uM) at 37ΰC for 15 min. In situ fluorescence was recorded using an Odyssey Li-Cor imaging platform.
2.9. Statistical methods
All bioassays were performed in triplicate. The arithmetic mean of a set of observed data was determined summation of the numerical values of each observation divided by the total number of observations. Error bars represent the standard deviation of the triplicate values relative to the arithmetic mean. Supervised hierarchical clustering by normalized expression of the UPS genes further indicated that a number of genes encoding catalytic, structural or regulatory subunits of the proteasome were upregulated in post-treatment samples. The ratio of individual gene expression in BMPCs isolated from HLA-sensitized patients post-treatment relative to that pre-treatment was calculated on an individual patient basis. A p-value cutoff of 0.05 was considered statistically significant (Fisher’s exact test). The ratio of gene products on western blots was compared using densitometric analysis on an Odyssey imaging platform. R version 3.5.0 was then used for paired tests to compare pre- vs. post-treatment blots by 2-sided analysis. In Fig. 6, given that n=8, a non-parametric Wilcoxon signed rank test was used.
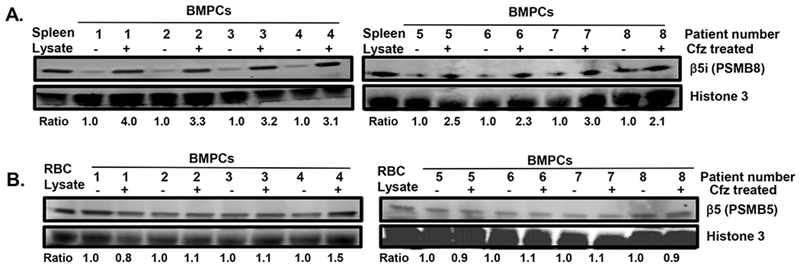
Figure 6.
Effect of in vitro carfilzomib treatment on β5i levels in BMPCs. Patient BMPCs (100,000 cells) were co-cultured with BMSC conditioned media for 72 h in the presence or absence of carfilzomib for 72 h. Cells were removed, pelleted and lysates prepared in 50 uL celLytic non-denaturing buffer. Histone 3 was used as a loading control. Lysates were separated on 4-12% bis-tris gels, transferred to PVDF membranes and probed using Abs to proteasome subunit β5i (PSMB8) (A) and β5 (PSMB5) (B). Erythrocytes (RBCs) contain constitutive 20S proteasomes and do not contain i20S proteasomes. RBC lysates was used as positive control to detect constitutive 20S proteasomes. Ratio indicates the relative intensity of a given proteasome subunit in lysates of that individual patient prior to and after carfilzomib treatment (N=8).
3. RESULTS
3.1. Co-culture with BMSC conditioned media extends the longevity of BMPCs
The short half-life of human BMPCs in vitro has significantly limited study of their growth and survival (30–33). Recent studies have shown that BMPCs cultured in vitro under well-defined conditions display a remarkable robustness, extended survival and produce immunoglobulins without contact with BM cells, if specific growth factors, e.g., APRIL (a proliferation-inducing ligand), IL-6, IL-12, and nutrients are provided. We and others have investigated the capacity of primary BM-derived stromal cells to maintain PC viability in vitro. PCs purified from BM died rapidly when plated in media, but a subpopulation of PCs survived and secreted high levels of Ab for up to 4 weeks when co-cultured with stromal cells (33). Importantly, the longevity of BMPCs from HLA-sensitized patients was extended upon co-culture with BMSC conditioned media supplemented with APRIL (200ng/mL) and IL-6 (10ng/mL). CD138+ cell survival was >50% at 6 days when cultured in BMSC conditioned media while the majority of CD138+ cells cultured in RPMI media alone were no longer viable at 3 days (Supp. Fig. 4).
3.2. BMPCs secrete HLA Abs that are reflective of profiles in patient serum
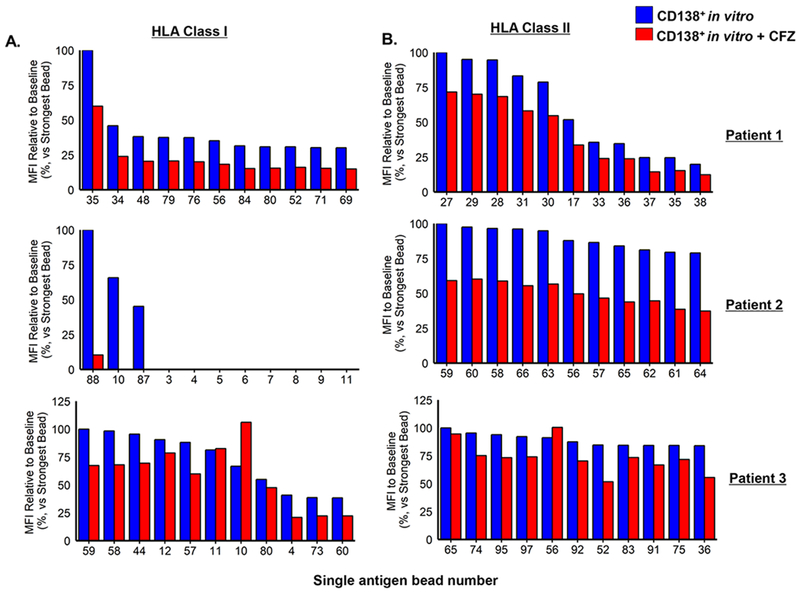
We then sought to determine whether CD138+ cells were a major source of HLA Ab production and whether these cells produced class I and II HLA Ab profiles similar to those present in the serum of HLA-sensitized patients. BM biopsies were obtained from HLA-sensitized patients, the BM mononuclear cells isolated and the CD138+ cells separated from the CD138- cells by positive selection. The BM mononuclear fraction, CD138+ and CD138- PCs were then co-cultured with BMSC conditioned media for 72 h. Culture supernatants were removed and the levels of class I and II HLA Abs in the supernatants determined. Representative SAB assay histograms from an individual HLA-sensitized patient indicated that the CD138+ BMPCs, but not the CD138- cells, produced class I and II Abs with similar hierarchical specificities to that seen in serum from the same patient (Fig. 1). Similar results were obtained using serum and cell fractions from two other HLA-sensitized patients to indicate that the CD138+ BMPCs are a major source of class I and II HLA Abs (Supp. Fig. 5A, B). Plots then generated to demonstrate that Ab levels in CD138+ culture supernatants correlated with the serum levels of HLA class I and class II Abs from the same three patients (Supp. Fig. 6A and 6B).
Figure 1.
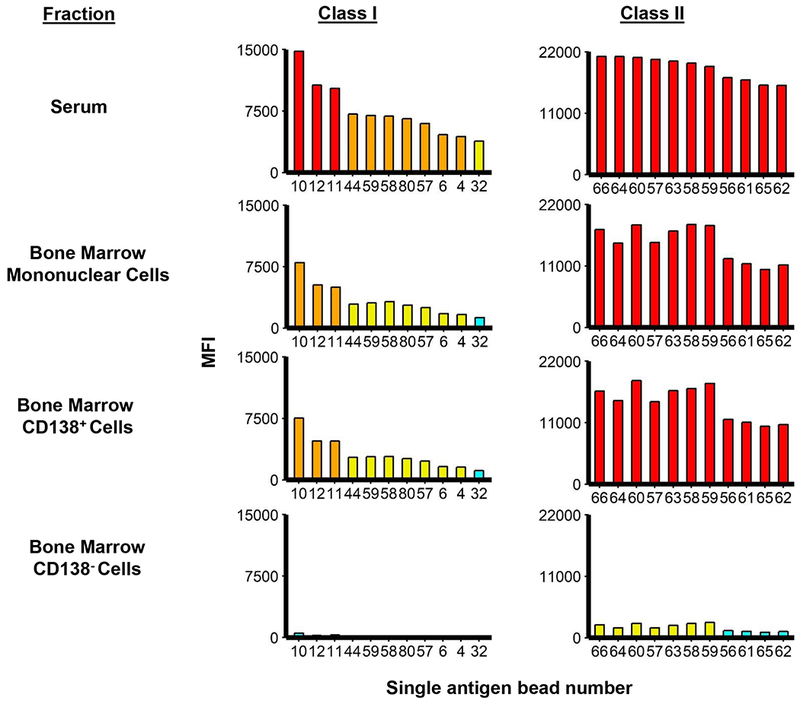
CD138+ BMPCs from HLA-sensitized patients produce class I and II Abs at levels comparable to those detected in patient serum. Class I and II Ab levels were detected, identified and titered in serum from HLA-sensitized patients and culture supernatants from BM cell fractions using the single antigen bead (SAB) assay (LABScreen®; One Lambda, Canoga Park, CA) assay on a Luminex platform (Austin, TX). BM mononuclear cells (20,000/assay), CD138+ (100,000/assay) and CD138− PCs (100,000/assay) from that same patient were incubated in 100 uL BMSC conditioned media supplemented with APRIL and IL-6 at 37°C for 72 h. Serum and cell culture supernatants were removed and 20 uL was used for SAB assays (48, 49). A representative histogram of the Luminex™ SAB results to determine the levels of class I and II HLA in patient serum and cell fractions is shown. Similar results were observed using serum and cells from two additional HLA-sensitized patients (see Supp. Figure 5A, B).
3.3. RNA-seq reveals proteasome genes upregulated in BMPCs after carfilzomib therapy
We initially sought to determine whether in vivo carfilzomib therapy reduced the relative number of BMPCs present in patient BM biopsies. The total number of live PCs recovered from patient BM was determined using trypan cell exclusion prior to and after in vivo carfilzomib. We detected a profound depletion in the percent of viable PCs recovered from patient BM, relative to the total number of nucleated cells recovered in that BM, following carfilzomib treatment. In vivo carfilzomib therapy depleted 60-70% of the BMPCs in HLA-sensitized patients (Fig. 2A).
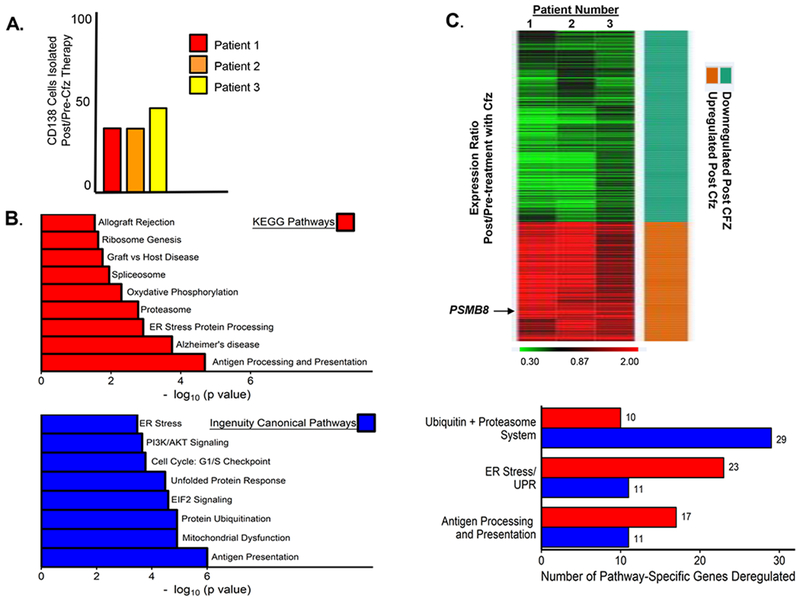
Figure 2.
Effect of in vivo carfilzomib therapy on gene expression in patient BMPCs. A. Proportion of live CD138+ cells recovered as a function of total bone marrow nucleated cells prior to and after in vivo carfilzomib therapy. Shown is the relative percent of live PCs recovered from the BM biopsies of three individual patients. Cell viability was determined using the trypan blue exclusion assay. Live PCs and the total number of nucleated BMMF cells isolated from individual patient BM biopsy were determined prior to and after in vivo carfilzomib therapy. Shown is the ratio of live CD138+PCs recovered from each patient post/pre-carfilzomib treatment. B. Unsupervised and supervised pathway analyses were performed to compare the genes differentially expressed in CD138+ PCs isolated from patients pre- and post in vivo carfilzomib therapy. KEGG and Ingenuity canonical pathway analyses were performed to identify the most significantly deregulated genes and pathways between BMPCs isolated pre- and post-carfilzomib therapy. Shown are the number of genes determined by either KEGG or Ingenuity canonical pathway analyses to be significantly deregulated in patient BMPCs following in vivo carfilzomib therapy. KEGG analysis generated an in-depth understanding of the differentially-expressed genes at the metabolic and functional levels, gene ontology functional annotation and KEGG pathway enrichment analysis were performed using DAVID (Database for Annotation, Visualization and Integration Discovery) http://david.abcc.ncifcrf.gov/. Canonical pathway analysis was performed using the Ingenuity Pathway Analysis tool (IPA, Ingenuity® Systems, www.ingenuity.com) employed using the DEGs. A p-value cutoff of 0.05 was considered statistically significant (Fisher’s exact test). C. Heat map depicting the genes down- or upregulated in each of the three HLA-sensitized patients following in vivo carfilzomib therapy. BMPCs were isolated from patients prior to and after in vivo carfilzomib therapy and then analyzed by RNA-seq. The heat map indicates genes that demonstrated a fold change ([Post]/[Pre]) greater than 1.5 in all three patients. Green indicates genes down regulated and red indicates those upregulated. A p-value cutoff of 0.05 was considered statistically significant (Fisher’s exact test).
We also sought to identify genes involved in BMPC survival following in vivo carfilzomib therapy. RNA-seq analysis was performed to detect genes significantly deregulated in CD138+ BMPCs isolated from HLA-sensitized patients after in vivo carfilzomib therapy relative to their expression level prior to therapy. A total of 2,024 differentially expressed transcripts that corresponded to 1,241 genes (1043 upregulated, 198 down regulated) were identified. Following in vivo therapy with carfilzomib, we found that the ten most upregulated genes in BMPCs were: MUC17, ZNF645, VN1R1, JAKMIP3, SH3TC2, MUC4, MAPT-IT1, SMCO3, NMNAT2, and MSL3P1 and the ten most down regulated genes were SATL1, PYGM, RPRM, ARHGEF25, SYNC, ABO, NPIPA2, SH3RF1, MROH6, and OR1J4 (Supp. Table 2).
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Ingenuity Canonical Pathway (IPA) analyses of differentially expressed genes implicated a number of molecular pathways and components that were upregulated in CD138+ BMPCs that survived carfilzomib therapy (Fig. 2B). Notably, these included a significant number of genes within the ubiquitin-proteasome system (UPS) differentially expressed with statistical significance between pre-treatment samples relative to post-treatment samples (Fig. 2C). We focused on genes within the UPS since this pathway is well known to undergo substantial perturbations following proteasome inhibition. The ten most up or down regulated genes within the UPS are shown in Supp. Table 3 and the ten most up- or down regulated proteasome/immunoproteasome genes are shown in Supp. Table 4. The ratio of individual gene expression in BMPCs isolated from HLA-sensitized patients post-treatment relative to that pre-treatment was calculated on an individual patient basis. The analysis revealed that PSMB8, which encodes the β5i catalytic subunit in i20S proteasomes, was the most significantly upregulated proteasome-associated gene in BMPCs isolated post-carfilzomib therapy (Fig. 2C). In addition, the FDA-approved PIs bortezomib, carfilzomib and ixazomib also demonstrate inhibitory effects on multiple catalytic subunits in addition to the proteasome β5 catalytic subunit encoded by PSMB5. PSMB8 is a particularly attractive target since it also encodes an immunoproteasome catalytic subunit that is also inhibited by carfilzomib. PSME1 and PSME2, which encode the proteasome activator PA28α/β subunits, were also upregulated following carfilzomib therapy (Supp. Fig. 7).
We also looked at the expression of known PC survival factors as well as cytokine and chemokine expression. Importantly, the expression of the APRIL, IL-6 and BAFF receptors was not significantly changed in BMPCs isolated following in vivo carfilzomib. Interestingly, a number of cytokine and chemokines and receptors were deregulated in BMPCs after carfilzomib, regardless of whether they are known to substantially affect PC survival (Supp. Table 5).
3.4. β5i and PA28 are upregulated in BMPCs upon in vivo carfilzomib therapy
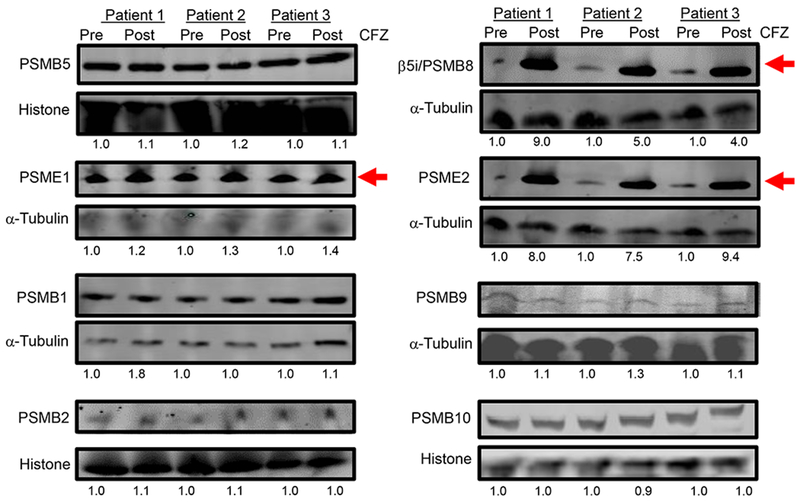
To extend the aforementioned transcriptomic findings, lysates were prepared from BMPCs isolated from individual HLA-sensitized patients before and after in vivo carfilzomib therapy. Western blotting indicated that β5i (PSMB8), PA28α and PA28β were significantly upregulated in BMPC lysates after carfilzomib therapy, relative to that observed prior to therapy, while other proteasome catalytic subunits β1, β1i, β2, β2i, and β5 were not significantly changed (Fig. 3).
Figure 3.
Effect of in vivo carfilzomib therapy on proteasomal catalytic and activator subunits in patient BMPCs. Western blot of c20S proteasome and i20S proteasome catalytic subunits and the PA28 activator subunits (PSME1 and PSME2) in BMPC lysates from HLA-sensitized patients isolated before or after in vivo carfilzomib therapy. Ratio represents the relative intensity of a band in lysates isolated after carfilzomib therapy relative to the loading control. Results shown are from three individual patients.
3.5. β5i incorporation into catalytically active i20S proteasomes upon carfilzomib therapy
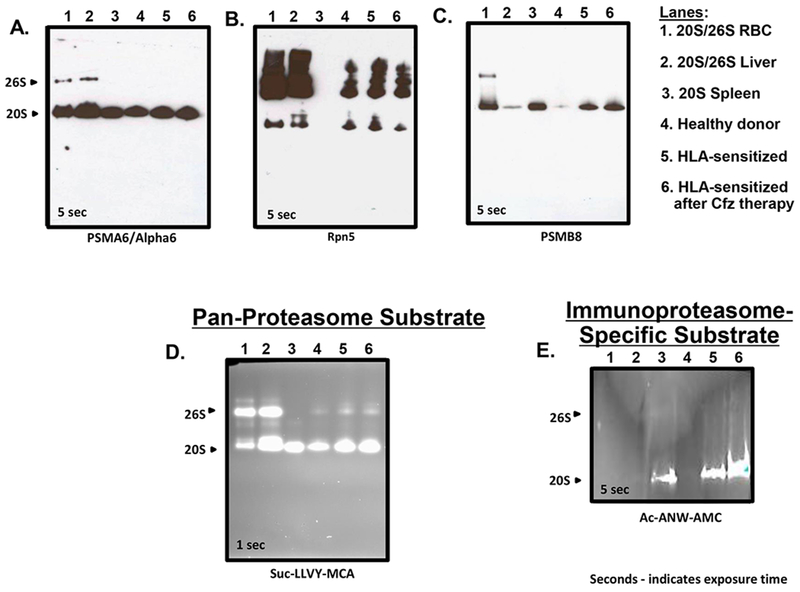
We next sought to determine whether PSMB8 upregulation upon carfilzomib therapy resulted in increased incorporation of the β5i subunit into i20S proteasomes. Proteasomes were affinity-purified from the BMPCs of healthy individuals and HLA-sensitized patients either prior to or after carfilzomib therapy and separated by native gel electrophoresis (33–35). Western blots were normalized using the proteasome structural subunit PSMA6 (Fig. 4A) and Rpn5 was used to detect 19S RPs in proteasome preparations from erythrocytes, liver and BMPC samples (Fig. 4B). Spleen has the greatest level of i20S proteasome expression compared to other organs and 20S spleen proteasomes were nearly exclusively i20S proteasomes lacking 19S RPs. Western blots indicated that β5i levels were greater in HLA-sensitized dialysis patient BMPC proteasome preparations and elevated further in preparations from BMPCs that survived in vivo carfilzomib therapy (Fig. 4C).
Figure 4.
Native gel electrophoresis to detect structural and functional changes within proteasomes isolated from patient BMPCs. A. Samples were run on 3-8% Bis-Tris protein gels (Invitrogen) for 45 V at 4°C overnight and subjected to western blotting using alpha6 (clone MCP20). Western blot using a proteasome α6 subunit-specific Ab against proteasomes isolated from human erythrocytes, liver or spleen (lanes 1-3). Samples from the PCs of a healthy adult (lane 4) or BMPCs from a single HLA-sensitized patient prior to and after in vivo carfilzomib therapy (lanes 5 and 6). Samples (5ug each) were loaded onto native gels and electrophoresed as described above. B. Western blot using a proteasome Rpn5 subunit-specific Ab with the same samples electrophoresed as above. C. Western blot using a proteasome β5i (PSMB8) subunit-specific Ab with the same samples electrophoresed as above. D. Activity overlay assay using the pan-proteasome fluorogenic substrate Suc-LLVY-MCA. E. Activity overlay assay using the i20S proteasome substrate Ac-ANW-AMC. Shown are representative western blots and overlay assays observed using lysates from a single patients. Similar results were observed using BMPCs isolated from two additional HLA-sensitized patient (Supp. Fig. 8).
3.6. i20S proteasome activity is increased in BMPCs upon in vivo carfilzomib therapy
The HLA sensitized patients treated with carfilzomib also differed from healthy controls in that they were on dialysis. Therefore, we also evaluated proteasome catalytic activity in healthy controls in addition to HLA-sensitized dialysis patients before and after in vivo carfilzomib therapy. Proteasomes were isolated from the CD138+ BMPCs of a single patient, separated by native gel electrophoresis and incubated with the pan-proteasome Ch-T-L substrate Suc-LLVY-MCA (Fig. 4D). Results indicated that Ch-T-L activity was increased in 20S proteasomes from the BMPCs of HLA-sensitized patients compared to BMPCs from healthy adults and further increased in proteasomes from BMPCs that survived carfilzomib therapy. Ac-ANW-AMC is a fluorogenic substrate preferentially cleaved by i20S proteasomes and is not hydrolyzed by c20s proteasomes (36). Ac-ANW-AMC hydrolysis by proteasomes isolated from healthy donor BMPCs was low but was significantly increased in proteasomes isolated from HLA-sensitized patient CD138+ BMPCs (Fig. 4E). Ac-ANW-AMC hydrolysis by proteasomes from BMPCs of carfilzomib treated HLA-sensitized patients was even greater. Results indicated that β5i was indeed incorporated into i20S proteasomes and there was a shift toward i20S proteasome-specific activities within the proteasome pool of CD138+ BMPCs that survived carfilzomib. Similar results were obtained using BMPCs from two additional HLA-sensitized patients (Supp. Fig. 8).
3.7. Carfilzomib reduces production of class I and II Abs by BMPCs cultured in vitro
Patient BMPCs were cultured in vitro with BMSC media either alone or in the presence of carfilzomib. Following 72 h of culture, the supernatants were removed and used for SAB assays (Fig. 5A, B). Results indicated that carfilzomib treatment in vitro reduced the level of most individual class I and II HLA Abs generated in the culture supernatants by 60-70%.
Figure 5.
Effect of carfilzomib treatment in vitro on the production of class I (A.) and II (B.) Abs by BMPCs from HLA-sensitized patients. Patient BMPCs (100,000 cells) were co-cultured in 100uL BMSC conditioned media for 72 h. Cell suspensions were centrifuged for 3 min at 3,000 rpm, culture supernatants removed and individual class I and II Ab levels measured using the SAB assay. Culture media was incubated alone as a negative control. Serum isolated from the whole blood of HLA-sensitized patients was used to measure the levels of individual class I and II Abs.
3.8. Carfilzomib treatment in vitro induces an increase of β5i (PSMB8) in BMPCs
CD138+ BMPCs were isolated from eight individual HLA-sensitized patients, co-cultured with BMSC conditioned media, and either left untreated or treated with carfilzomib. Lysates were prepared and analyzed by western blotting to determine the effect of carfilzomib on the levels of the proteasome β5 and β5i subunits. Western blotting indicated a significant increase in β5i in lysates from the patients that were analyzed (Fig. 6A). In contrast, little change was observed in the β5 subunit (Fig. 6B).
3.9. BMPCs that survive carfilzomib therapy are sensitive to immunoproteasome inhibition
BMPCs that had been isolated from three patients prior to and after in vivo carfilzomib therapy were treated with individual PIs and the number of apoptotic cells then measured using annexin V-FITC (Fig. 7). BMPCs isolated from patients prior to carfilzomib therapy were relatively sensitive to the proteasome inhibitors bortezomib, carfilzomib and ixazomib. In contrast, BMPCs isolated after in vivo carfilzomib therapy were more sensitive to ONX-0914 (Fig. 7).
Figure 7.
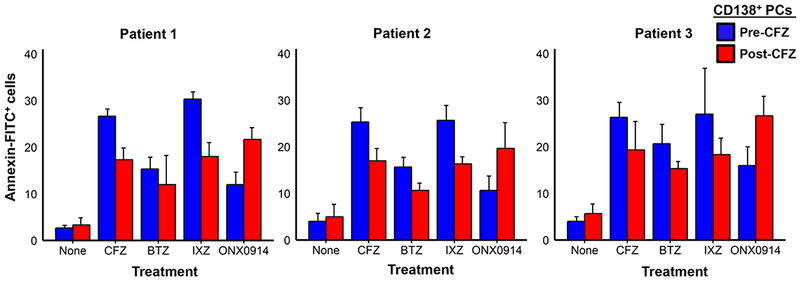
Effect of different PIs on BMPCs isolated before and after in vivo carfilzomib therapy. Cells were incubated with the PIs bortezomib, carfilzomib or ixazomib (10 nM) or ONX-0914 (100 nM) for 16h and then stained to detect apoptotic cells using the annexin V-FITC apoptosis detection kit. Shown are representative results from three HLA-sensitized patients.
DISCUSSION
Major progress has been made in understanding PC biology over the past decade, and more recently with respect to elucidating transcriptional regulatory networks that underlie B cell differentiation and PC generation in immune responses. Our prior experience in treating Ab-mediated rejection and in desensitization in transplantation, has led us to realize that plasmablasts are exquisitely sensitive to bortezomib therapy, whereas BMPCs demonstrate considerable, yet partial resistance. This realization led us to begin to explore the basis for the heterogeneity within BMPC populations following PI therapy. We reasoned that understanding drug resistance would enable development of more effective targeting of BMPCs. This rationale underpinned the present study. A key first step in the study was to determine whether BMPCs represented a major source of HLA Abs, as suggested in the existing literature. The similarities in HLA Ab level hierarchy between patient serum and that produced in vitro by BMPCs supported the concept that long-term HLA Ab production is derived from BMPCs, thereby supporting our selection of this cellular population to focus the present studies.
RNA-seq and biochemical analyses revealed that the β5i subunit, β5i-specific catalytic activity and i20S proteasomes were substantially upregulated in BMPCs isolated from HLA-sensitized dialysis patients following carfilzomib therapy. Transcriptomic studies of BMPCs before and after in vivo carfilzomib treatment revealed an unforeseen plasticity of the UPS and proteasomes within Ab-producing BMPCs. Unexpectedly, we found that i20S proteasomes are upregulated in response to carfilzomib, and were characterized by both upregulation of both β5i catalytic activity and also PA28 activator subunit expression (PSME1 and PSME2). These observations suggest that specific targeting of β5i activity, and also potentially preventing PA28 association with the core 20S particles, represent potential avenues for future PI-base therapies that enhancing BMPC depletion. These studies are unique in that they investigate for the first time, the effects of in vivo carfilzomib treatment on human BMPCs and outline a unique adaptive mechanism that may limit clinical effectiveness of PI therapy.
An important question that we began to explore was whether PI resistance was predetermined, i.e., that specific BMPC subpopulations existed prior to therapy that would demonstrate PI resistance, or whether it arose as a result of BMPC adaptation upon drug treatment. Interestingly, in vitro treatment of cultured BMPCs for 24 h induced responses similar to those observed following in vivo treatment, indicative of an adaptive response. The results are consistent with the known reduced sensitivity of i20S proteasomes to agents that target the c20S proteasome. Modulation of proteasome catalytic subunit gene expression suggests proteasomal adaptation as a new chemoresistance mechanism that promotes the survival of BMPCs. β5i alterations were also reliably accompanied by PA28 alterations. While the function of PA28 is not fully elucidated, association with i20S proteasomes suggests that it may also potentially contribute to chemoresistance.
i20S proteasomes have been proposed as a potential new target for suppressing autoimmune and inflammatory processes (37–43). Importantly, the β5i-selective inhibitor ONX0914 has shown to suppress autoreactive immune responses in mouse models of arthritis and diabetes (44). Furthermore, β5i inhibition prevented experimental colitis (45), Hashimoto’s thyroiditis (46) and graft-versus-host disease in an MHC-matched minor histocompatibility antigen-disparate murine model (47). In addition to providing irreversible proteasome inhibition, carfilzomib displays a distinct, advantageous adverse-event profile compared to bortezomib for the treatment of MM patients. Understanding the mechanisms of resistance to proteasome inhibition will not only allow better use of PIs, but may also facilitate rational design of more effective synergistic drug combinations for clinical trials to reduce the BMPCs responsible for alloantibody production. Alternatively, the present study suggests that serial treatment with constitutive followed by immunoproteasome-specific inhibitors may provide an effective alternative to concomitant/ simultaneous therapy. While elimination of autoreactive PCs by c20S PIs represents a viable strategy for Ab-mediated diseases, our study reveals that targeting i20S proteasomes may further enhance PC depletion and abrogate HLA Ab production.
Our studies suggest that the development of effective long-term desensitization treatments will require genomic and functional studies to uncover mechanisms of chemoresistance. As shown here, bulk RNA-transcriptomic analyses revealed alterations in the UPS that occur during the initiation and progression of HLA-sensitization and/or upon carfilzomib therapy, and this knowledge can now be exploited for molecular diagnostics and the development of novel desensitization therapies. In so doing, we highlight the value of clinical trials that integrate genomic analyses to guide the use of currently available drugs and enable development of new targeted therapeutics.
Supplementary Material
ACKNOWLEDGEMENTS.
Funding: ESW received research funds from Amgen Pharmaceuticals, Inc, Thousand Oaks, CA. Funding also provided from the National Institute of Allergy and Infectious Diseases of the NIH: 1R01AI139141-01A1 and 1R56AI139141-01.
ABBREVIATIONS:
- DSA
Donor-specific antibodies
- BMPCs
bone marrow plasma cells
- Abs
antibodies
- PIs
proteasome inhibitors
- MM
multiple myeloma
- ESRD
end stage renal disease
- HLA
human leukocyte antigen
- AMR
antibody-mediated rejection
- IVIg
intravenous immunoglobulin
- FDA
Federal Drug Administration
- IRB
Institutional Review Board
- NCT
National Clinical Trial
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- IPA
Ingenuity Canonical Pathway Analysis
- UPS
ubiquitin-proteasome system
- SAB
single antigen bead
Footnotes
DISCLOSURE. The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
DATA SHARING. The data that support the findings will be available following an embargo from the date of publication to allow for commercialization of research findings.
REFERENCES
- 1.Stewart AK, Rajkumar SV, Dimopoulos MA et al. for the ASPIRE Investigators. Carfilzomib, lenalidomide, and dexamethasone for relapsed multiple myeloma. N. Engl. J. Med 2015; 372: 142–152. [DOI] [PubMed] [Google Scholar]
- 2.Richardson PG, Sonneveld P, Schuster MW et al. for the Assessment of Proteasome Inhibition for Extending Remissions (APEX) Investigators. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med 2005; 352:2487–2498. [DOI] [PubMed] [Google Scholar]
- 3.Moreau P, Masszi T, Grzasko N et al. for the TOURMALINE-MM1 Study Group. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N. Engl. J. Med 2016; 374:1621–1634. [DOI] [PubMed] [Google Scholar]
- 4.Everly MJ, Everly JJ, Susskind B et al. Bortezomib provides effective therapy for antibody- and cell-mediated acute rejection. Transplantation 2008; 86:(12):1754–1761. [DOI] [PubMed] [Google Scholar]
- 5.Abecassis M, Bartlett ST, Collins AJ et al. Kidney transplantation as primary therapy for end-stage renal disease: A National Kidney Foundation/Kidney Disease Outcomes Quality Initiative (NKF/KDOQITM) conference. Clin J Am Soc Nephrol 2018; 3:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grams ME, Massie AB, Coresh J, Segev DL. Trends in the timing of pre-emptive kidney transplantation. J Am Soc Nephrol 2015; 22:1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishbane S, Nair V. Opportunities for Increasing the Rate of Preemptive Kidney Transplantation. Clin J Am Soc Nephrol 2018; 13:(8)1280–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin WM III, Valujskikh A, Fairchild RL. Mechanisms of antibody-mediated acute and chronic rejection of kidney allografts. Curr Opin Organ Transplant 2016; 21:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery RA, Loupy A, Segev DL. Antibody-mediated rejection: new approaches in prevention and management. Am J Transplant 2018; 18 (Suppl. 3):3–17. [DOI] [PubMed] [Google Scholar]
- 10.Garces JC, Giusti S, Staffeld-Coit C et al. Antibody-mediated rejection: a review. Ochsner J. 2017; 17:46–55. [PMC free article] [PubMed] [Google Scholar]
- 11.Reed EF, Demetris AJ, Hammond E et al. , Acute antibody-mediated rejection of cardiac transplants. J. Heart Lung Transplant. 2006; 25:153–159. [DOI] [PubMed] [Google Scholar]
- 12.de Kort H, Roufosse C, Bajema IM et al. , Pancreas transplantation, antibodies and rejection: where do we stand? Curr. Opin. Organ. Transplant 2013; 18:337–344. [DOI] [PubMed] [Google Scholar]
- 13.Slifka MK, Ahmed R, Long-term antibody production is sustained by antibody-secreting cells in the bone marrow following acute viral infection. Ann N Y Acad Sci 1996; 797:166–176. [DOI] [PubMed] [Google Scholar]
- 14.Martin F, Chan AC, Pathogenic roles of B cells in human autoimmunity: insights from the clinic. Immunity 2004; 20:517–527. [DOI] [PubMed] [Google Scholar]
- 15.Uchida J, Machida T, Iwai T. et al. Desensitization protocol in highly HLA-sensitized and ABO-incompatible high titer kidney transplantation. Transplant Proc. 2010; 42:3998–4002. [DOI] [PubMed] [Google Scholar]
- 16.Ramos EJ, Pollinger HS, Stegall,. The effect of desensitization protocols on human splenic B-cell populations in vivo. Am. J. Transplant 2007; 7(2):402–407. [DOI] [PubMed] [Google Scholar]
- 17.Vo AA, Lukovsky M, Toyoda M. et al. , Rituximab and intravenous immune globulin for desensitization during renal transplantation. N. Engl. J. Med 2008; 359:242–251. [DOI] [PubMed] [Google Scholar]
- 18.Sethi S, Choi J, Toyoda M, Desensitization: Overcoming the Immunologic Barriers to Transplantation. J. Immunol. Res 2017; 2017:6804678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cenci S, Oliva L, Cerruti F et al. , Pivotal advance: protein synthesis modulates responsiveness of differentiating and malignant plasma cells to proteasome inhibitors J. Leukoc. Biol 2012; 92:921–931. [DOI] [PubMed] [Google Scholar]
- 20.Chauhan D, Hideshima T, Mitsiades C, et al. , Proteasome inhibitor therapy in multiple myeloma. Mol. Cancer Ther 2005; 4:686–692. [DOI] [PubMed] [Google Scholar]
- 21.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem 2009; 78:477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloetzel P-M Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol 2001; 2:179–188. [DOI] [PubMed] [Google Scholar]
- 23.Ebstein F, Kloetzel P-M, Kruger E, et al. , Emerging roles of immunoproteasomes beyond MHC class I antigen processing. Cell. Mol. Life. 2012; 69:2543–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremblay S, Shields A, Alloway RR et al. , A Prospective Carfilzomib-Based Desensitization Trial: Phase 1 Results. Am. J. Transplant 2016; 16:(suppl 3). [Google Scholar]
- 25.Woodle ES, Shields R, Ejaz NS et al. , Prospective iterative trial of proteasome inhibitor-based desensitization. Am. J. Transplant 2015; 15(1):101–118. [DOI] [PubMed] [Google Scholar]
- 26.Fairfax KA, Kallies A, Nutt SL, Plasma cell development: From B-cell subsets to long-term survival niches. Seminars Immunol. 2008; 20:49–58. [DOI] [PubMed] [Google Scholar]
- 27.Perry DK, Burns JM, Pollinger HS et al. , Proteasome inhibition causes apoptosis of normal human plasma cells preventing alloantibody production. Am J Transplant 2009; 9:201–209. [DOI] [PubMed] [Google Scholar]
- 28.Diwan TS, Raghavaiah S, Burns JM et al. , The impact of proteasome inhibition on alloantibody-producing plasma cells in vivo. Transplantation 2011; 91:536–541. [DOI] [PubMed] [Google Scholar]
- 29.Moravec RA, O’Brien MA, Daily WJ, et al. , Cell-Based bioluminescent assays for all three proteasome activities in a homogeneous format. Anal. Biochem 2009; 387:294–302. [DOI] [PubMed] [Google Scholar]
- 30.Cocco M, Stephenson MA, Care D et al. In vitro generation of long-lived human plasma cells. J Immunol. 2012; 189(12):5773–5785. [DOI] [PubMed] [Google Scholar]
- 31.Jourdan M, Caraux A, De Vos J et al. , An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood 2014;114:5173–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jourdan M, Cren M, Robert N et al. , IL-6 supports the generation of human long-lived plasma cells in combination with either APRIL or stromal cell-soluble factors. Leukemia 2014; 28(8): 1647–1656. [DOI] [PubMed] [Google Scholar]
- 33.Minges Wols HA, Underhill GH, Kansas GS, et al. , The role of bone marrow-derived stromal cells in the maintenance of plasma cell longevity. J. Immunol 2002; 169:4213–4221. [DOI] [PubMed] [Google Scholar]
- 34.Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005; 398: 353–363. [DOI] [PubMed] [Google Scholar]
- 35.Lönnroth I, Oshalim M, Lange S et al. , Interaction of proteasomes and complement C3, assay of antisecretory factor in blood. J. Immunoassay Immunochem. 2016; 37:43–54. [DOI] [PubMed] [Google Scholar]
- 36.Karreci ES, Fan H, Uehara M et al. Brief treatment with a highly selective immunoproteasome inhibitor promotes long-term cardiac allograft acceptance in mice. Proc Natl Acad Sci U.S.A 2016; 113(52):E8425–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal AK, Xing C, De Martino GN. PSMB8 encoding the β5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am. J. Hum. Genet 2010; 87:866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura A, Maekawa Y, Uehara H et al. , A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Invest 2011;121: 10:4150–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arima K, Kinoshita A, Mishima H et al. , Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Nat. Acad. Sci. U.S.A 2011; 108:14914–14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torrelo A, Patel S, Colmenero I et al. , Chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) syndrome. J. Am. Acad. Dermatol 2010; 62: 489–495. [DOI] [PubMed] [Google Scholar]
- 41.Brehm A, Liu Y, Sheikh A et al. , Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest. 2015; 125:4196–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraile A, Nieto A, Vinasco J et al. , Association of large molecular weight proteasome 7 gene polymorphism with ankylosing spondylitis. Arthritis Rheum. 1998; 41:560–562. [DOI] [PubMed] [Google Scholar]
- 43.Deng GY,Muir A, Maclaren NK, et al. Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: population and family studies. Am. J. Hum. Genet 1995; 56(2):528–534. [PMC free article] [PubMed] [Google Scholar]
- 44.Muchamuel T, Basler M, Aujay MA et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med 2009; 15:781–787. [DOI] [PubMed] [Google Scholar]
- 45.Basler M, Dajee M, Moll C et al. , Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol. 2010; 185:634–641. [DOI] [PubMed] [Google Scholar]
- 46.Nagayama Y, Nakahara M, Shimamura M et al. Prophylactic and therapeutic efficacies of a selective inhibitor of the immunoproteasome for Hashimoto’s thyroiditis, but not for Graves’ hyperthyroidism, in mice. Clin Exp Immunol. 2012; 168:268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silberberg J, Matos J,Dziopa E et al. , Inhibition of the immunoproteasome subunit LMP7 with ONX0914 ameliorates graft-versus-host disease in an MHC-matched minor histocompatibility antigen-disparate murine model. Biol Blood Marrow Transplant 2015; 21(9): 1555–1564. [DOI] [PubMed] [Google Scholar]
- 48.Walsh RC, Brailey P, Girnita A. et al. , Early and late acute antibody-mediated rejection differ immunologically and in response to proteasome inhibition. Transplantation 2011; 91: 1218–1226. [DOI] [PubMed] [Google Scholar]
- 49.Reed EF, Rao P, Zhang Z et al. , Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA. Am J Transplant 2013; 13: 1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.