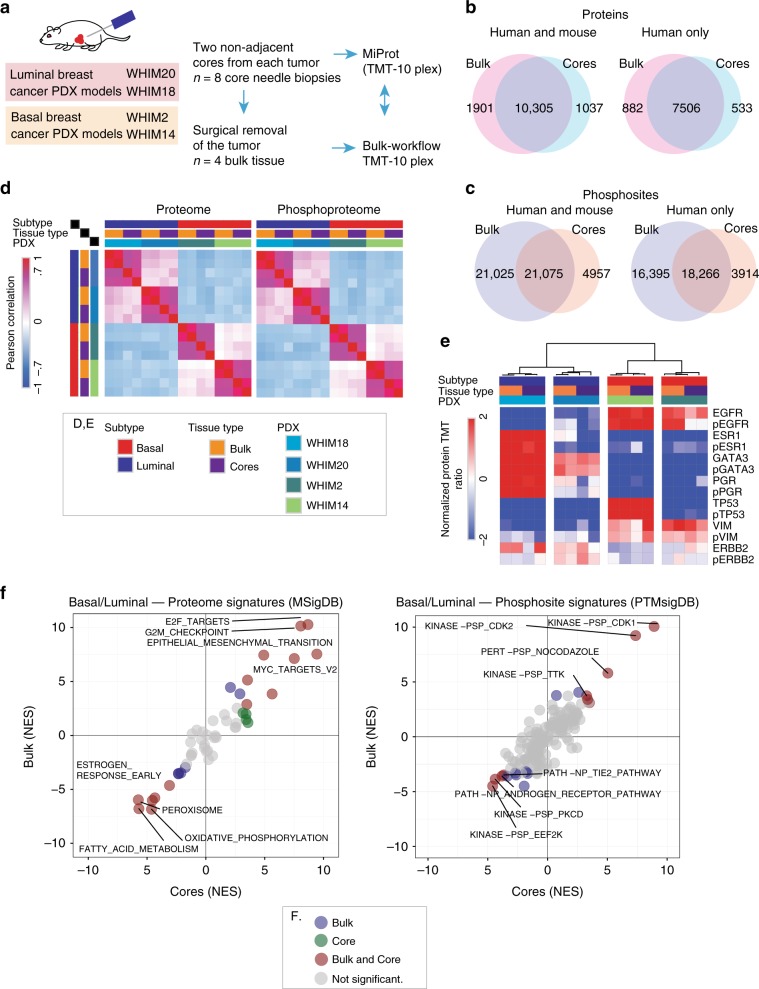
Fig. 2. Evaluation of the BioText and MiProt workflow on preclinical PDX models.
a Non-adjacent, core needle biopsies were collected from 2 basal and 2 luminal PDX models followed by surgical removal of tumors. Proteomic and phosphoproteomic characterization of cores was performed using the MiProt workflow, and the bulk tissue was characterized using the CPTAC workflow described in Mertins et al8. b Venn-diagram shows the number of overlap between human and mouse or human proteins quantified in cores and bulk tissue. c Venn-diagram shows the overlap between human and mouse or human phosphosites. d Pearson correlation of TMT ratios for proteins (left) and phosphosites (right) between each sample from both cores and bulk across all 4 PDX models. e The heatmap shows the TMT ratios for key differentially regulated Luminal vs. Basal breast cancer associated proteins and phosphoproteins (average expression of identified phosphosites) identified across both bulk and cores experiments. f Gene-centric and phosphosite-centric pathway or kinase activity enrichment analysis was performed using GSEA (MSigDB “Cancer Hallmarks”, left) and PTM-SEA (PTMSigDB, right), respectively, for Luminal-Basal differences captured in bulk (y-axis) and core (x-axis) tissue. limma derived signed Log10 p-values were used to pre-rank differential features for both GSEA and PTM-SEA analysis. The pathway/phospho-signatures that are significant in both cores and bulk are indicated in brown.