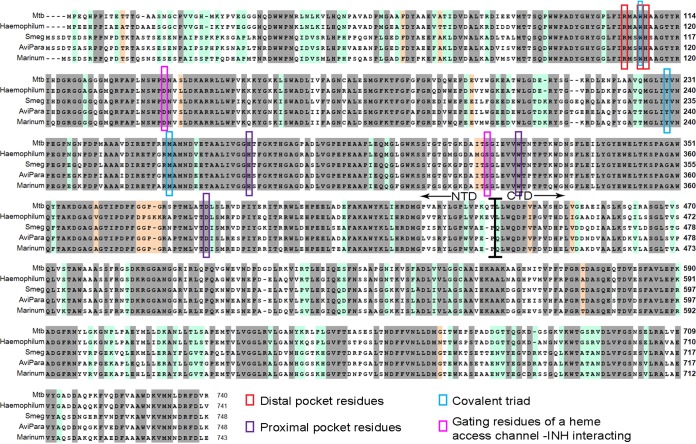
FIG 4.
Multiple sequence alignment of KatG proteins from M. tuberculosis, M. haemophilum, M. marinum, M. avium subsp. paratuberculosis, M. smegmatis and M. avium subsp. avium. Identical residues, highlighted in gray, comprise ∼65% of the residues, and additional ∼20% identity in M. tuberculosis /Haemophilum versus Avp/Mari/Smeg highlighted in light green. Important residues in the heme distal pocket (red), heme proximal pocket (purple), covalent triad (blue) and gating residues of the heme access channel important for the interaction with INH (pink) are all marked. The border between the N- and C-terminal domains is also shown, around residue 450.