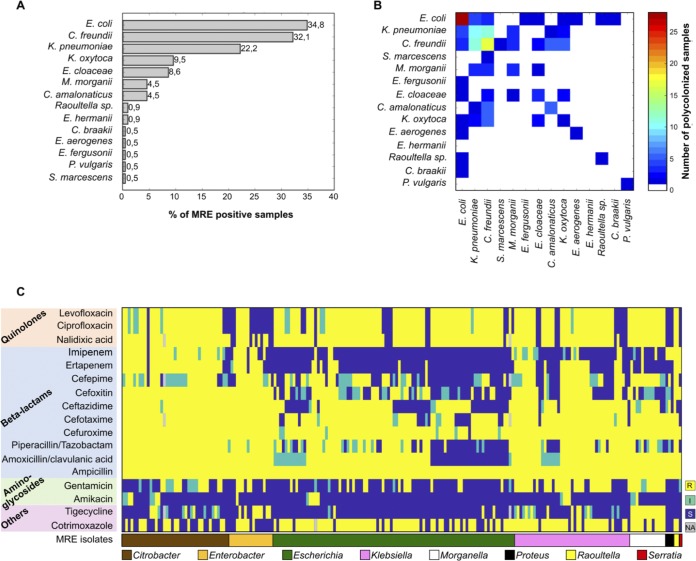
FIG 1.
MRE-positive samples show diverse compositions and antibiotic resistance patterns of detected MRE isolates. MRE were detected in 221 samples collected from 80 of the analyzed patients. (A) The most frequently isolated MRE belonged to the species Escherichia coli. Other detected MRE in order of prevalence were Citrobacter freundii, Klebsiella pneumoniae, Klebsiella oxytoca, Enterobacter cloacae, Morganella morganii, Citrobacter amalonaticus, Raoultella spp., and Escherichia hermannii. Other MRE species, including Citrobacter braakii, Proteus vulgaris, Serratia marcescens, Enterobacter aerogenes, and Escherichia fergusonii, were detected in only one sample. (B) In 45 of the 221 MRE-positive samples (20.4%), MRE belonging to more than one bacterial species were identified, and in 70 of the MRE-positive samples (31.7%), MRE strains belonging to the same species but with different antibiotic resistance pattern were identified. (C) Antibiotic resistance patterns of MRE isolates detected in 221 positive samples. When two isolates from the same sample had exactly the same resistance patterns and taxonomies, only one of the two isolates was shown. Columns and rows were grouped based on MRE taxonomy and antibiotics’ class. S, susceptible; I, intermediate resistance phenotype; R, resistant. NA, not analyzed.