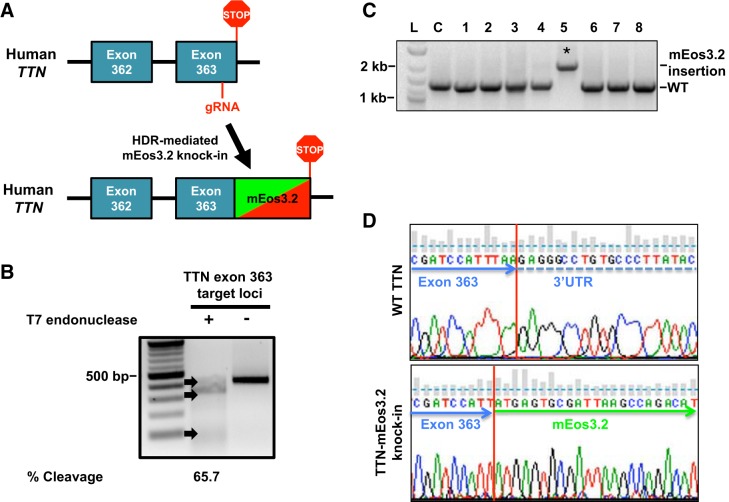
Fig. 1.
Generation of titin-mEos3.2 knock-in model in human induced pluripotent stem cells (hiPSCs). A: titin-mEos3.2 knock-in strategy to replace the stop codon in titin’s terminal exon 363 with an in frame mEos3.2 knock-in into titin’s terminal exon 363. A guide (g)RNA targeting titin exon 363 for cleavage was coelectroporated with a homology directed repair template (mEos3.2, with 600-bp titin flanking homology arms) in hiPSCs. B: gel image of T7 endonuclease assay demonstrates titin-specific gRNA targets exon 363 for cleavage in HEK 293 cells. Briefly, HEK 293 were transiently transfected with 1 μg of PX 459 plasmid encoding a titin-specific gRNA and Cas9 for 36 h. HEK 293 cells were then subjected to antibiotic selection (500 ng/mL, puromycin) for 48 h. HEK 293 cells were then harvested for DNA extraction. A 421-bp loci where the double-stranded breaks occurred was PCR amplified. The PCR product was denatured and reannealed before being subjected to T7 endonuclease. Lane 1: 100-bp ladder. Lane 2: treated with T7 endonuclease. Lane 3: not treated. DNA was visualized by ethidium bromide (EtBr) staining and %cleavage {[sum of cleaved band intensities/(total band intensities)] × 100} was quantified by band densitometry. C: gel image of mEos3.2 sequence integration in hiPSC. hiPSC were electroporated with 10 μg of gRNA and 500 ng of titin-mEos3.2 HDR template. Individual colonies hiPSC were screened for mEos3.2 insertion by PCR using primers flanking outside of the repair template. PCR products were ran on a 1% agarose gel and stained with EtBr. Lane L: 100-bp ladder. Lane C: control. Lanes 1–8: individual hiPSC clones. *mEos3.2 sequence integration. D: chromatogram of genomic sequences from hiPSC with and without mEos3.2 sequence integration. WT, wild type.