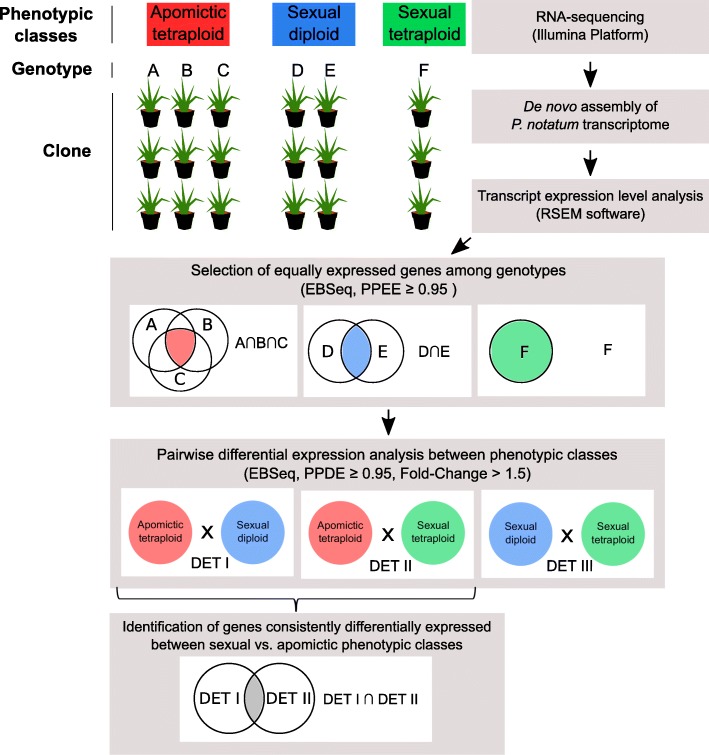
Fig. 6.
Workflow of differential expression analyses used in this study. We performed pairwise differential expression analyses of all three phenotypic classes (apomictic tetraploid “4X apo”, sexual diploid “2X sex” and sexual tetraploid “4X sex”). Each phenotypic class had different genotypes with three clones. First, the objective was to select a list of equally expressed transcripts among genotypes belonging to the same phenotypic class. EBSeq was used to estimate the pairwise posterior probabilities of a transcript being equally expressed (PPEE) among genotypes of the same phenotypic class as follows: apomictic tetraploid (A × B; A × C; B × C) and sexual diploid (D × E). Transcripts with PPEE ≥0.95 were kept for the second step of the analysis. Second, another round of pairwise differential expression analyses among phenotypic classes was performed as follows: (i) 4X apo vs. 2X sex; (ii) 4X apo vs. 4X sex; and (iii) 2X sex vs. 4X sex. The objective was to select a list of transcripts that presented pairwise posterior probabilities of being differentially expressed (PPDE ≥0.95). Additionally, we identified a list of transcripts that were consistently differentially expressed between the sexual and apomictic samples independently of the ploidy level (diploid or tetraploid)