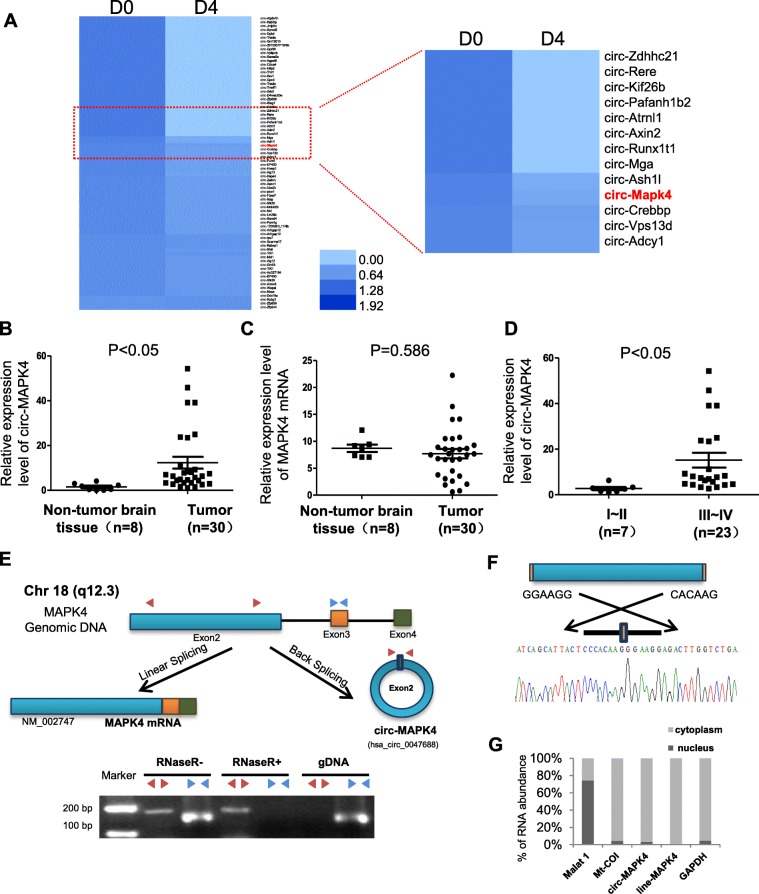
Fig. 1.
Characterization of circ-MAPK4 in human neural embryoid body and gliomas. (a) Heat map of microarray data which identified aberrantly expressed circRNAs in the first day (D0) and the 4th day (D4) upon inducing neural differentiation of murine P19 embryonic carcinoma (EC). (b) circ-MAPK4 is upregulated in glioma tissues compared with normal brain tissues (P < 0.05). (c) The level of MAPK4 mRNA did not show significant difference between glioma tissues and normal brain tissues (P > 0.05). (d) circ-MAPK4 is highly expressed in gliomas patients with advanced stages III + IV compared with I + II (P < 0.05). (e) Upper panel: Schematic representation of circ-MAPK4 (hsa_circ_0047688) formation. Divergent (red) and convergent (blue) primers were designed to amplify the back-splicing and linear products. Lower panel: Compared to the MAPK4 mRNA, RT-PCR revealed that circ-MAPK4 resistant to the RNase R, which also remain 80–90% after digested by RNase R. No product was amplified to genomic DNA using divergent primers. (f) The back-splice junction site of circ-MAPK4 was validated by RT–PCR followed by Sanger sequencing. (g) By the Nucleocytoplasmic separation experiment, circ-MAPK4 was identified in the cytoplasm mainly, which is consistent with the MAPK4 mRNA. MALAT and Mt-Coi is the marker of nucleus and cytoplasm, respectively. GAPDH was used as control