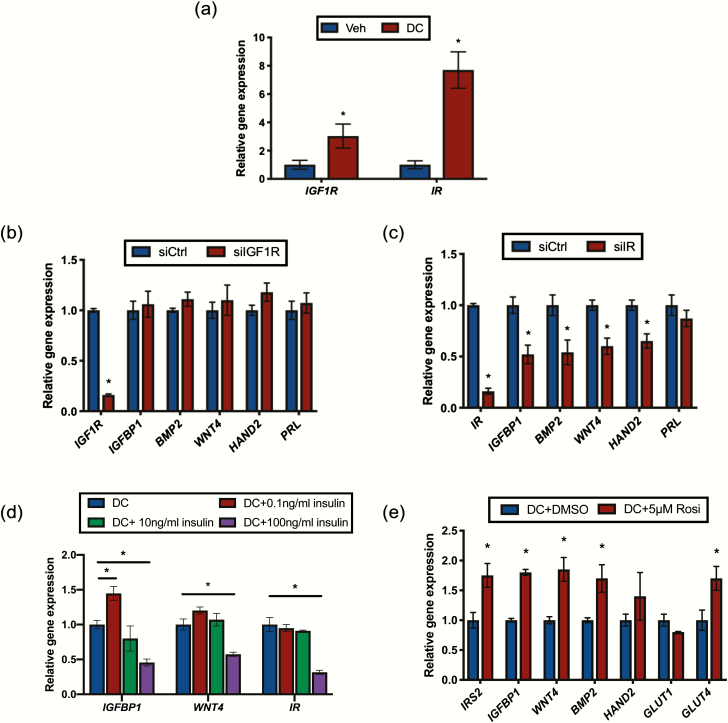
Figure 3.
IRS2 acts downstream of the insulin receptor (IR) to regulate decidualization. (A) Gene expression analysis was performed with RNA isolated from HESC treated with vehicle (ethanol) or DC for 72 hours using primers specific to IGF1R and IR. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to vehicle. Following siRNA mediated knockdown of IGF1R(B) or IR(C) and stimulation with DC for 72 hours, gene expression analysis was performed using primers for IGFBP1, BMP2, WNT4, HAND2, and PRL. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to siCtrl. (D) HESC were co-stimulated with DC and the indicated concentration of insulin for 72 hours. Gene expression analysis was performed using primers for IGFBP1, WNT4, and IR. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to DC alone. (E) Gene expression analysis was performed with RNA isolated from HESC treated with DC with or without 5μM rosiglitazone (Rosi) for 72 hours using primers specific to IRS2, IGFBP1, BMP2, WNT4, HAND2, GLUT1, and GLUT4. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to DC alone.