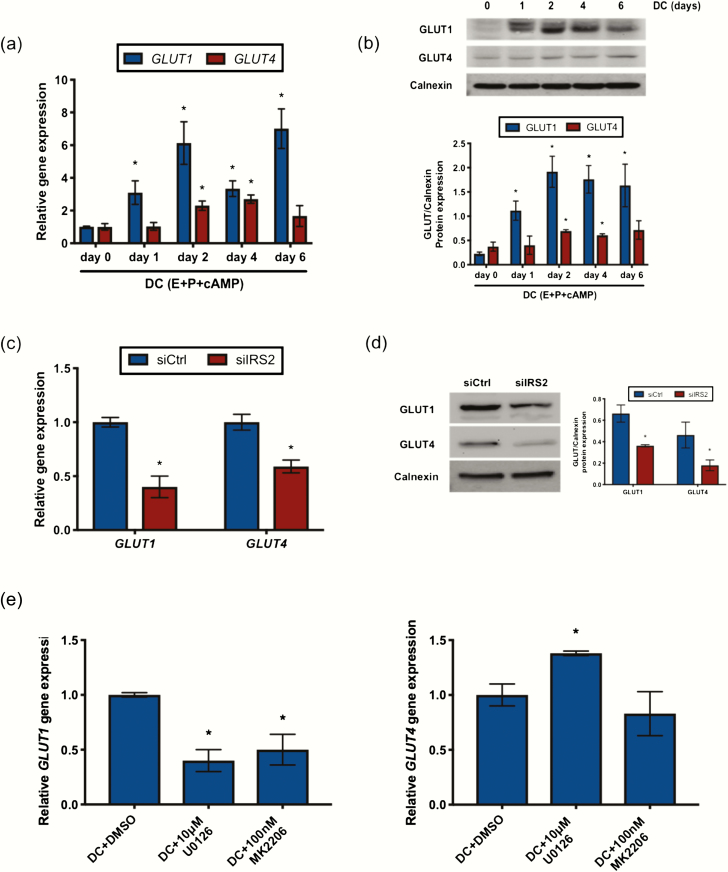
Figure 5.
IRS2 regulates GLUT expression during decidualization. GLUT mRNA (A) and protein (B) expression was observed in HESC treated with DC for 0 to 6 days. (A) Gene expression analysis was performed using primers specific for GLUT1 and GLUT4. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to day 0. (B) Whole cell lysates were analyzed by Western blot and probed with antibodies specific to GLUT1 and GLUT4. Calnexin is provided as a loading control. Densitometry analysis is provided (below). GLUT mRNA (C) and protein (D) expression was observed in HESC stimulated for 72 hours with DC following siRNA mediated knockdown of IRS2. (C) Gene expression analysis was performed using primers specific for GLUT1 and GLUT4. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to siCtrl. (D) Whole cell lysates were analyzed by Western blot and probed with antibodies specific for GLUT1 and GLUT4. Calnexin is provided as a loading control. Densitometry analysis is provided (right). (E) Gene expression analysis was performed with RNA isolated from HESC treated with DC and DMSO or either 10μM U0126 or 100nM MK2206 for 72 hours using primers specific for GLUT1 and GLUT4. 36B4 was used for normalization. Data are represented as the mean fold induction ± SEM. *P < 0.05 relative to DC+DMSO.