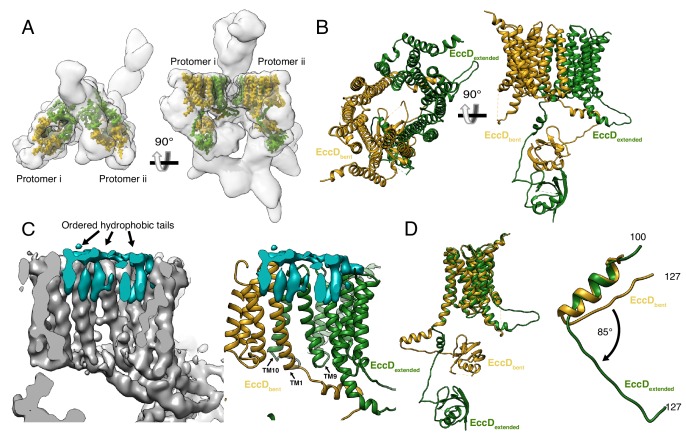
Figure 2. The structure of EccD3.
(A) EccD3-bent (yellow) and EccD3-extended (green) in the context of the overall ESX-3 dimer (gray transparency). (B) Atomic models of EccD3-bent and EccD3-extended (C) An unsharpened electron microscopy density map of the ESX-3 dimer shows extra densities consistent with lipid or detergent molecules (teal) on the periplasmic face of the EccD3 cavity. (D) EccD3-bent (yellow) and EccD3-extended (green) aligned based on the transmembrane regions shows two distinct conformations of the EccD3 cytoplasmic domains. Amino acids 100–127 of EccD3 adopt a bent (yellow) and an extended (green) conformation.