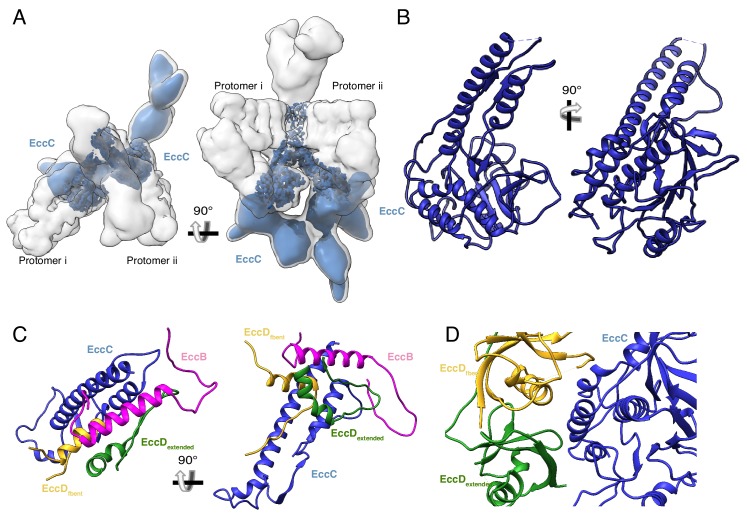
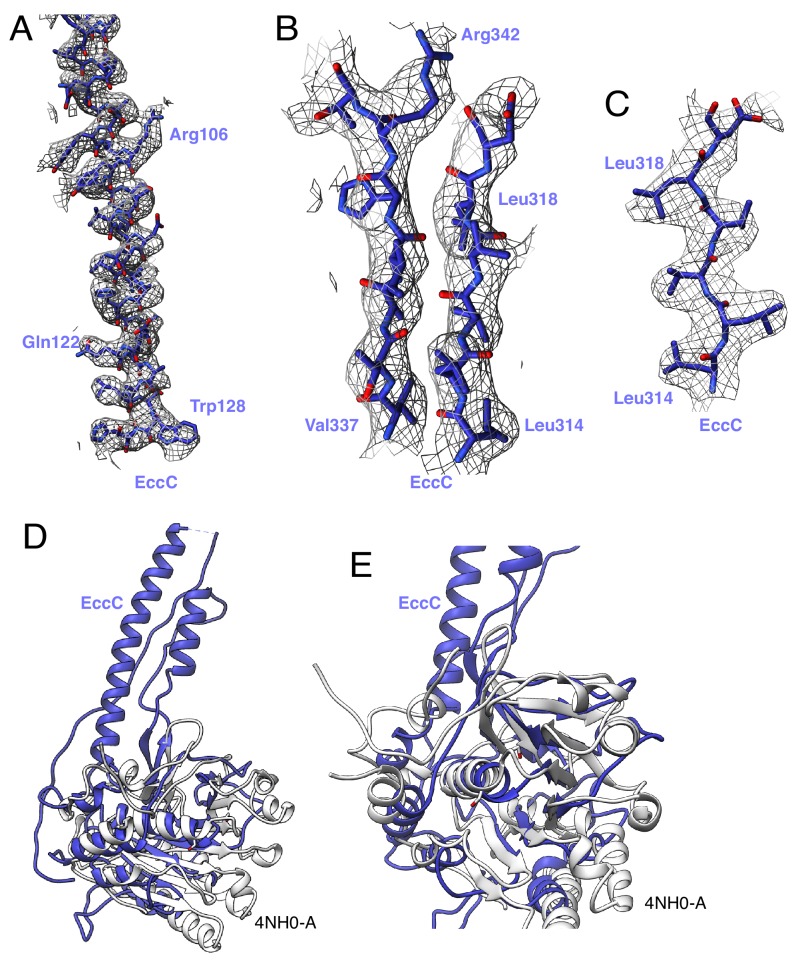
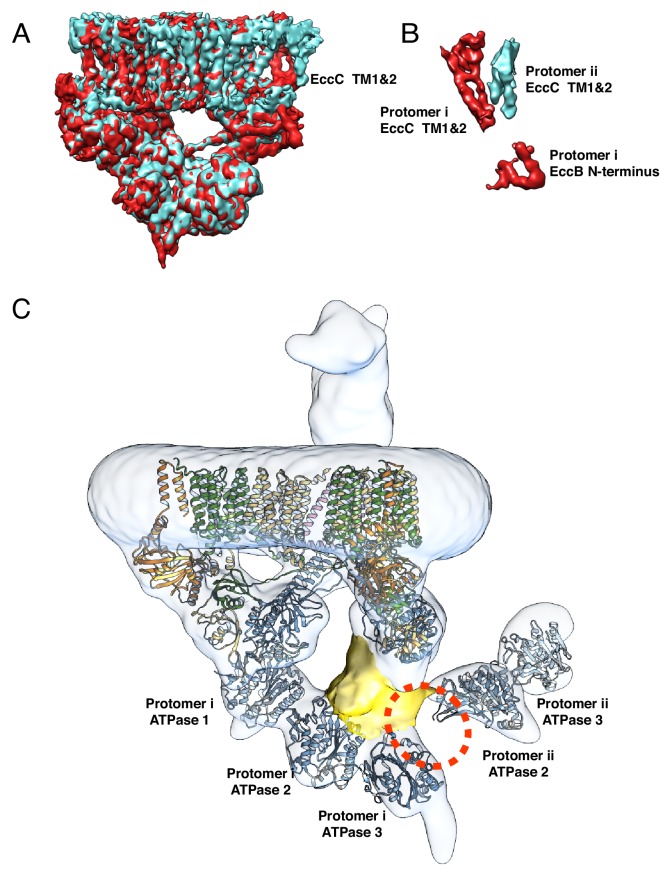
(A) Overlay of the focused refined maps of the transmembrane and upper cytoplasmic regions of protomer i (red) and protomer ii (blue). The two protomers are nearly identical except in the transmembrane domains of EccC3 and the N-terminal tail of EccB3. (B) Density subtraction of the two protomers in A highlights the regions of difference. Specifically, the transmembrane helixes of EccC3 adopt distinct conformations across protomers. (C) In protomer i, 4NH0, the crystal structure of EccC from T. curvata, fits well in the density. In protomer ii, ATPase 2 and 3 fit well into the density but the position of ATPase 1 in the crystal structure, shown by the red dotted circle, does not fit into the density. A density of about the right size for ATPase 1 is shown in yellow. Fitting of ATPase 1 in this density requires the disruption of the interface between ATPase 1 and ATPase 2.