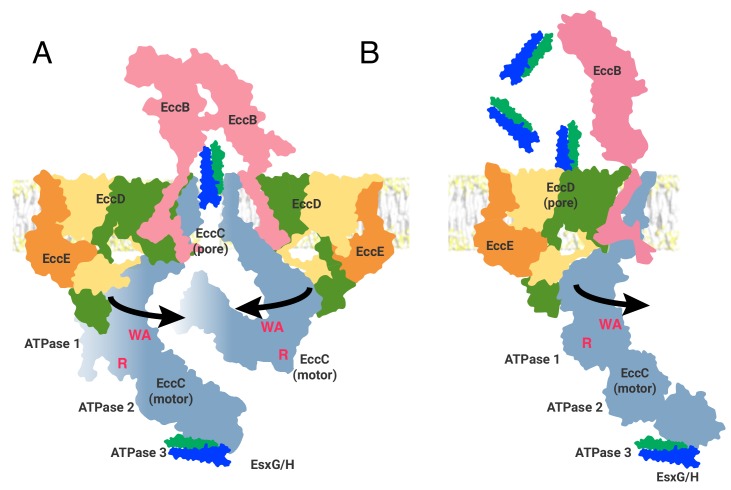
Figure 6. Two models of the ESX-3 translocon complex.
ATPase activity entails, at a minimum, oligomerization of ATPase 1 to bring the R-finger (R) into proximity of the catalytic site, marked by the Walker A motif (WA). This requires at least 65 Å of movement from the position seen in the structure. (A) The first model of ESX substrate secretion involves trimerization of the ESX-3 dimer followed by multimerization of the EccC ATPase domains into a stack of one to four rings of ATPases (B) The second model shown through the function of a single protomer of the ESX-3 complex. Substrates are selected by interaction with ATPase 3 of EccC and transported via the upper cytoplasmic region to the EccD cavity for secretion.