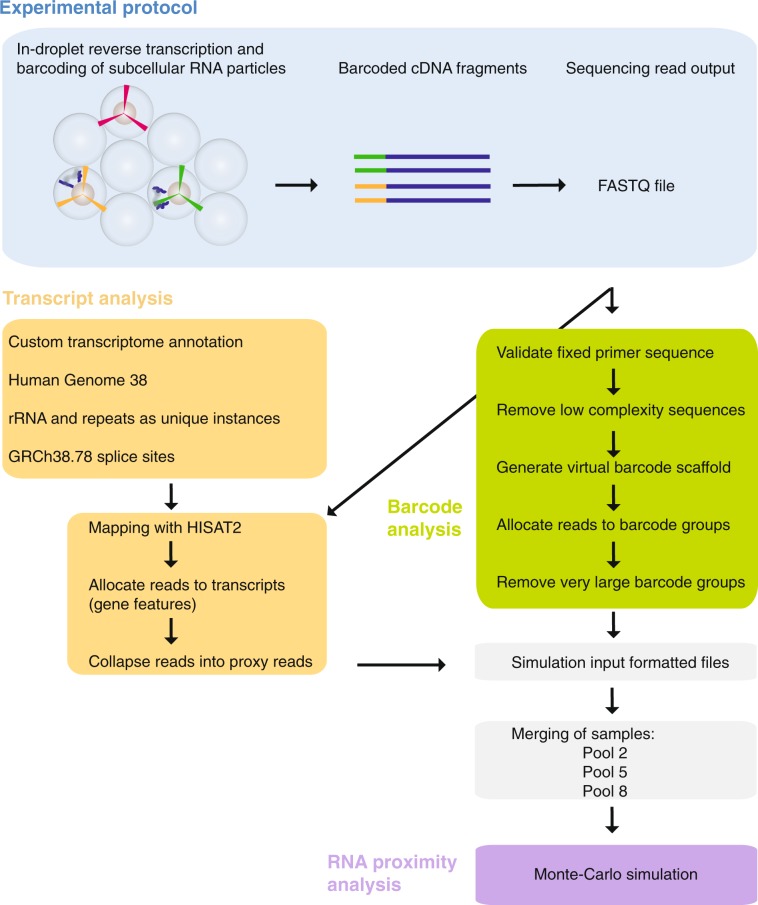
Fig. 1.
Workflow of Proximity RNA-seq, CloseCall and sample processing. Reverse transcription and RNA co-barcoding in droplets generate sequencing libraries with cDNAs, whose RNA templates were in spatial proximity, sharing the same barcode (blue box). After Illumina sequencing, single-end sequence reads from the FASTQ file are validated to contain a defined primer sequence. Barcodes of cDNAs are extracted and, if near-identical, grouped together (green box). In parallel, the cDNA part of each read is mapped onto the genome and allocated to a custom transcriptome annotation (yellow box). The Monte-Carlo simulation takes merged datasets as input and randomises cDNA – barcode pairings to derive expected co-barcoding frequencies for RNA pairs.