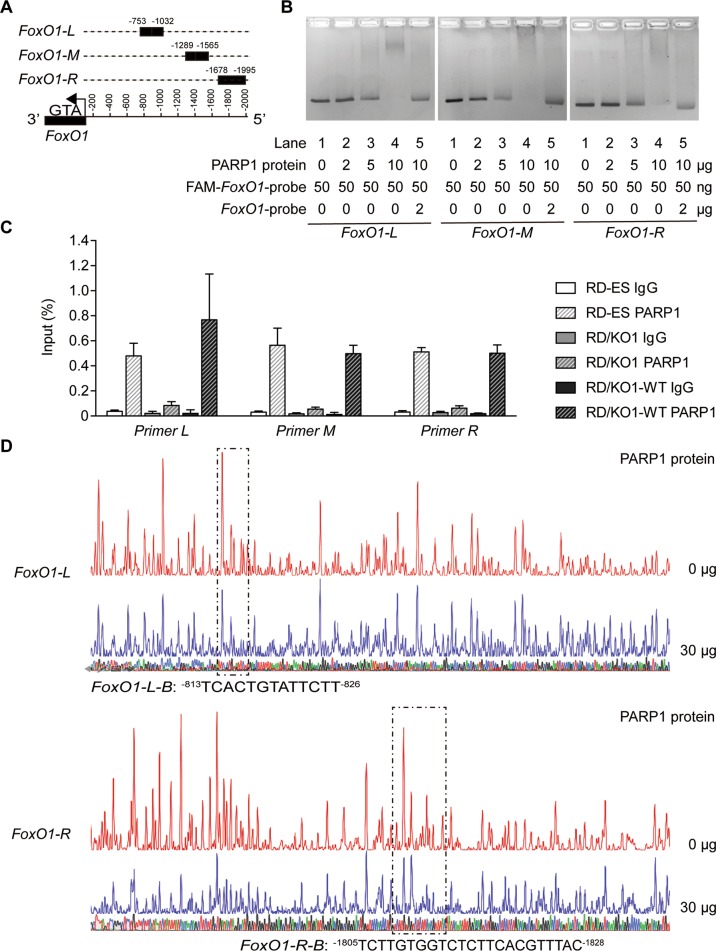
Fig. 3. PARP1 binding to particular regions on the FoxO1 promoter.
a Schematic representation of the locations of particular nucleotide fragments (FoxO1-L, -M, and -R) on the FoxO1 promoter analyzed by EMSA. b The binding of purified PARP1 to particular nucleotide fragments (FoxO1-L, -M, and -R) were analyzed by EMSA. For the competition assays, 40-fold excess of unlabeled DNA fragments were added to the reaction mixture before adding FAM-labeled probes, and the labeled PARP1-FoxO1 complexes were almost completely displaced. c RD-ES, RD/KO1, and RD/KO1-WT cells were subjected to ChIP analyses using the antibody against PARP1 and an isotype-matched IgG as a negative control. The association of PARP1 with the FoxO1 gene promoter was quantified by RT-qPCR using primers targeting FoxO1-L, -M, and -R, respectively. Error bars represent the SD. d Identification of PARP1-protected regions on FoxO1-L and FoxO1-R by DNase I footprinting assays. Electropherograms showed the whole region of the FoxO1-L/R after digestion with DNase I following incubation in the presence (blue) or absence (red) of PARP1. The DNA sequences of the PARP1-protected regions were marked with dashed rectangles and denoted as FoxO1-L-B (−813TCACTGTATTCTT−826) and FoxO1-R-B (−1805TCTTGTGGTCTCTTCACGTTTAC−1828).