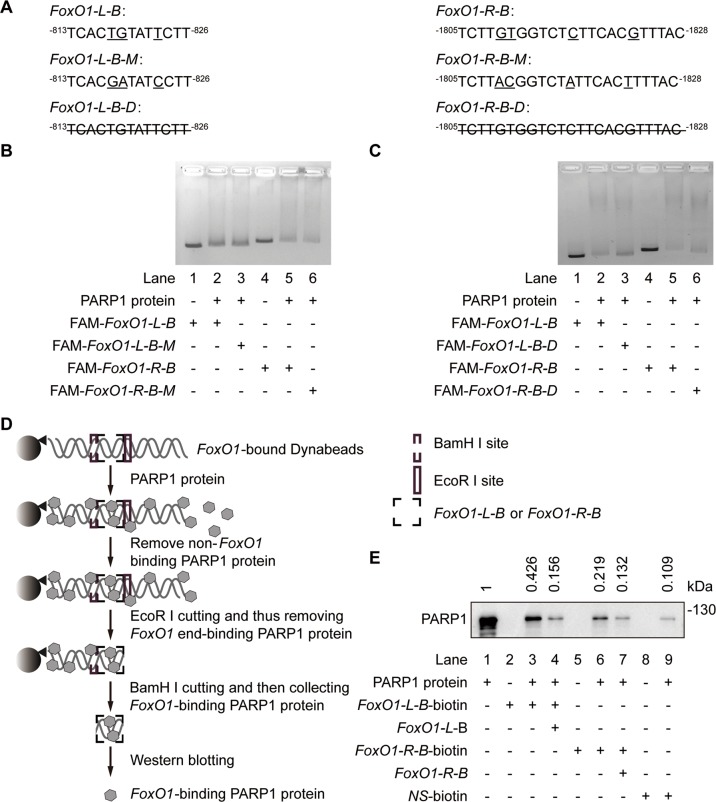
Fig. 4. PARP1 binding to the specific DNA sequences on the FoxO1 promoter.
a The FAM-labeled probes containing sequences of FoxO1-L-B (left) and FoxO1-R-B (right) and corresponding mutated probes (underlined; FoxO1-L-B-M and FoxO1-R-B-M), and the deleted sequences (line-through; FoxO1-L-B-D and FoxO1-R-B-D). b EMSA was carried out using normal or mutated FAM-labeled probes. The amount of DNA-protein complexes detected in FAM-labeled mutant probes was similar to that in FAM-labeled normal probes. c EMSA was carried out using FAM-labeled normal and deletion probes. Fewer DNA-protein complexes were detected with FAM-labeled deletion probes than the FAM-labeled normal probes followed by PARP1 incubation. d Schematic representation of the flanking restriction enhanced pulldown (FREP). A biotinylated DNA fragment is conjugated to streptavidin-coated magnetic Dynabeads (Invitrogen, Carlsbad, CA). This fragment is engineered to include the FoxO1-L-B or FoxO1-R-B specific (“bait”) sequence (black dashed box), flanked by restriction enzyme cleavage sites for BamH I proximally (gray dashed box) and EcoR I distally (gray box). DNA-beads are mixed with PARP1 protein. A free non-biotinylated FoxO1-L-B or FoxO1-R-B DNA fragment can be included in the control reaction at this stage as a specific competitor. Magnetic separation and wash remove non-DNA binding PARP1 protein. EcoR I digestion releases 3′ DNA end-binding PARP1, and BamH I digestion separates the sequence-specific FoxO1-L-B or FoxO1-R-B binding PARP1 from the 5′ DNA and Dynabeads. Western blotting identifies PARP1 binding to FoxO1-L-B or FoxO1-R-B. e The PARP1-DNA complexes cut with EcoR I and BamH I were detected by western blotting. The relative levels of FoxO1-bound PARP1 were presented as the ratio of FoxO1-bound PARP1 band intensity/PARP1 input band intensity when the value of PARP1 input band intensity was normalized as l. Lane 1: PARP1 inputs, lane 2: labeled FoxO1-L-B-beads, lane 3: PARP1 with labeled FoxO1-L-B-beads, lane 4: PARP1 with labeled FoxO1-L-B-beads and cold competitor (40-fold excess of free FoxO1-L-B-beads), lane 5: labeled FoxO1-R-B-beads, lane 6: PARP1 with labeled FoxO1-R-B-beads, lane 7: PARP1 with labeled FoxO1-R-B-beads and cold competitor (40-fold excess of free FoxO1-R-B-beads), lane 8: a labeled non-specific DNA sequence (NS-beads) and lane 9: PARP1 with labeled NS-beads.