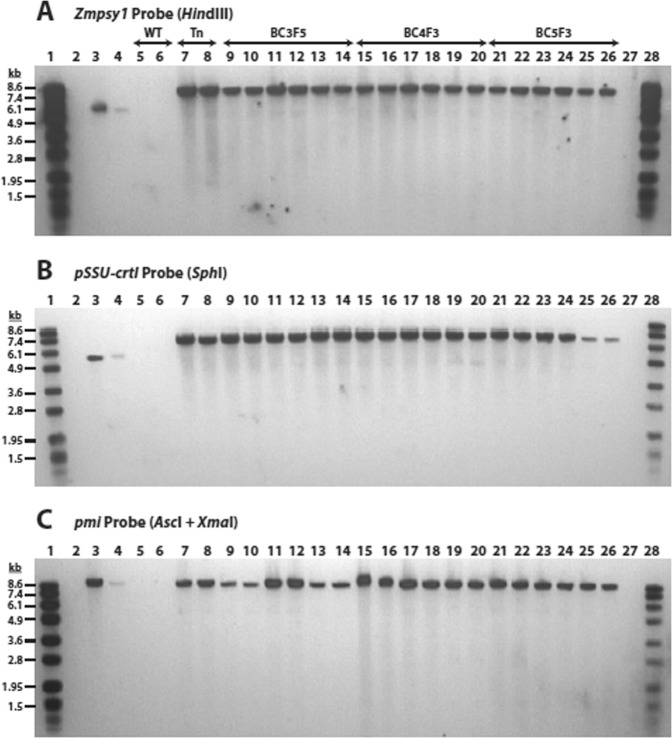
Figure 1.
Samples of DNA from individual plants of event GR2E in Kaybonnet (Tn; lanes 7–8, as replicates), BRRI dhan 29 (BC3F5, lanes 9–10; BC4F3, lanes 15–16; and BC5F3, lanes 21–22), IR64 (BC3F5, lanes 11–12; BC4F3, lanes 17–18; and BC5F3, lanes 23–24), and PSB Rc82 (BC3F5, lanes 13–14; BC4F3, lanes 19–20; and BC5F3, lanes 25–26) germplasm backgrounds and negative control DNA from Kaybonnet rice (lanes 5–6) were subjected to Southern blot analysis. For this, 5 μg genomic DNA was digested with HindIII (panel A), SphI (panel B), or AscI plus XmaI (panel C) followed by agarose gel electrophoresis and transfer onto nylon membrane. Positive control samples consisted of negative control Kaybonnet rice containing either one (lane 3) or 0.2 (lane 4) copy equivalents of pSYN12424 plasmid DNA digested with SphI (panels A and B) or AscI + XmaI (panel C), Blots were hybridized with DIG-labelled probes for Zmpsy1 (panel A), pSSU-crtI (panel B), or pmi (panel C). Following washing, hybridized probes and DIG-labelled molecular weight markers VII (lanes 1 and 28) were visualized using a chemiluminescent detection. Lanes 2 and 27 were blank on all gels (adapted from GR2E-FFP submitted study reports).