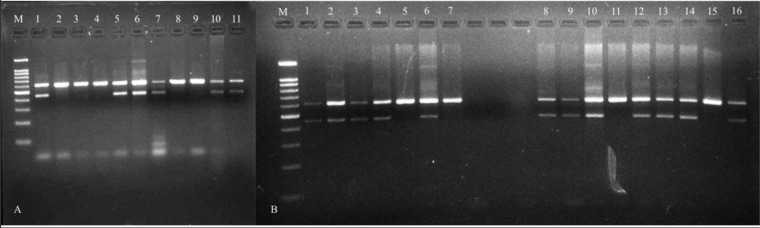
Figure 2.
(A) Standardization of modified ARMS PCR assay from the genomic DNA of Trichophyton mentagrophytes complex and Trichophyton rubrum; Lane 1 = Trichophyton rubrum (NCCPF 900041) with higher MICs to terbinafine having T1189C mutation; Lane 2, 3, 4 = Trichophyton mentagrophytes complex (NCCPF 800035, 800036, 800038) with low MICs to terbinafine. Lane 5, 6 = Trichophyton mentagrophytes complex (NCCPF 800022, 800023) with higher MIC’s to terbinafine having T1189C mutation; Lane 7,10,11 Trichophyton mentagrophytes complex (NCCPF 800062, 800063, 800064) with higher MICs to terbinafine having C1191A mutation; Lane 8, 9 = Trichophyton mentagrophytes complex (NCCPF 800035, 800036) with low MICs to terbinafine. Lane 1–6 were amplified using mutation primer-1 and lane 7–11 amplified using mutation primer-2. (M = 100 base pair ladder). (B) Validation of the modified ARMS PCR assay against genomic DNA of representative T. mentagrophytes complex with higher MICs to terbinafine. Lane 1, 2, 3, 4, 6 = Trichophyton mentagrophytes complex (NCCPF 800062, 800063, 800064, 800065, 800066) with high terbinafine MIC’s and C1191A mutation; lane 5, 7 = Trichophyton mentagrophytes complex (NCCPF 800043, 800044) with low MICs to terbinafine; Lane 8, 9, 10, 12, 13, 14, 16 = Trichophyton mentagrophytes complex (NCCPF 800055, 800056, 800052, 800053, 800060, 800061, 800057) with T1189C mutation; Lane 11, 15, = Trichophyton mentagrophytes complex (NCCPF 800043, 800044) with low terbinafine MICs. Lane 1–7 were amplified using mutation primer-2 and Lane 8–16 amplified with mutation primer-1. (M = 100 base pair ladder).