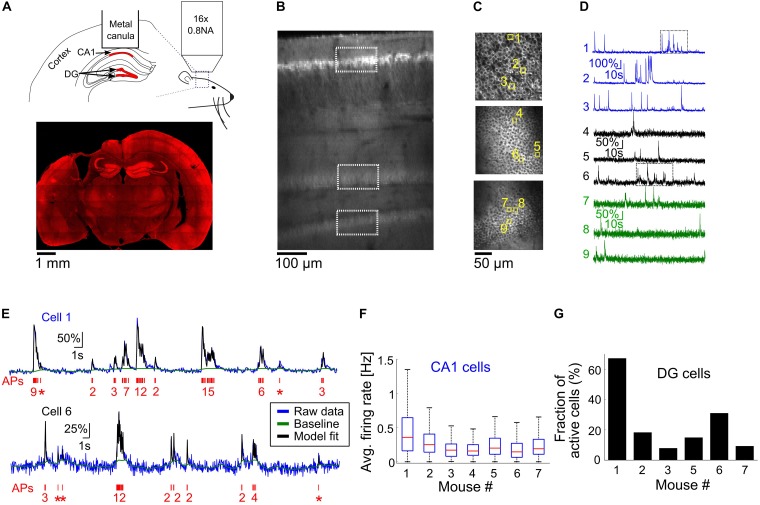
FIGURE 2.
Optical recording from mouse CA1 and DG regions of the hippocampus. (A) Top, schematic illustration of the mouse hippocampal recording experiments. We implanted metal cannulas with a glass bottom on top of the hippocampi of transgenic Thy-jRGECO1a mice to gain optical access to CA1 and DG neurons (shown in red). Hippocampus geometry was adopted from a mouse brain atlas (Dong, 2008). Bottom, a coronal section from the brain of mouse 1 shows the location of the hippocampal window and native jRGECO1a fluorescence signal throughout the brain. Note that some distortion to the brain tissue occurred when the metal cannula was detached from it. (B) In vivo dorsal-to-ventral view of a Thy1-jRGECO1a line 8.31 transgenic mouse expressing the red GECI jRGECO1a in the hippocampus 3 weeks after a craniotomy surgery. The tissue on top of the hippocampal formation was removed to expose the hippocampus and a z-stack series of two-photon microscopy images shows CA1 and DG regions. White rectangles indicate the location of the CA1 and DG (upper and lower blades) projection neurons, labeled by jRGECO1a. (C) Images of CA1 pyramidal cells (top image) and DG granular cells from the upper and lower blades of DG (middle and bottom images, respectively), corresponding to the respective rectangles shown in panel (B). (D) ΔF/F0 traces of the nine cells highlighted with yellow squares in panel (C). Note the differences in firing pattern and amplitude between the CA1 and DG cells. (E) Model fit (Deneux et al., 2016) (black line) to the raw fluorescence signal (blue line) of cells one and six [upper and lower traces, respectively, corresponding to the rectangles shown in panel (D)] to extract the baseline signal (green line) and AP firing (red, bottom, single AP events are represented with asterisks). The model was fed with jRGECO1a biophysical parameters (Dana et al., 2016), and showed good agreement to the recorded data from both CA1 (top) and DG (bottom) neurons. (F) Summary of average firing rates of all recorded CA1 pyramidal cells from seven mice before the initiation of cuprizone diet (328–623 cells from a single recording session for each mouse, 3,167 cells total). Red lines correspond to medians, blue boxes show the 25th–75th percentile range, whisker length is the shorter of 1.5 times the 25th–75th range or the extreme data point. Outliers are not shown. (G) Fraction of active DG neurons from all recorded DG neurons from six mice before the start of cuprizone diet (470–997 cells from a single recording session for each mouse, DG cells of mouse 4 were not visible, 3,113 cells total).