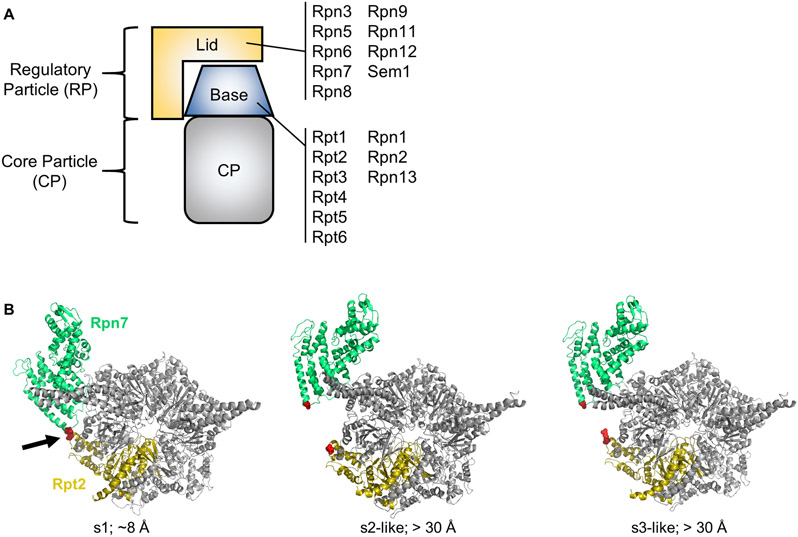
Figure 1.
A. Cartoon depiction of the 26S proteasome. The subunit composition of the lid and base subcomplexes is shown. B. Rpn7-D123 and Rpt2-R407 are juxtaposed in the s1 conformation of the yeast 26S proteasome (red spheres indicated by arrow) (PDB 4CR2), but are distant in the s2-s6 states. In the middle panel, the s2 state (PDB 4CR3) is shown, but the s5 state is highly similar. In the right panel, the s3 state (PDB 4CR4) is shown, but the s4 and s6 states are highly similar. The approximate distances in Angstroms between the Rpn7-D123 and Rpt2-R407 alpha carbons is shown. Panel B is modified from (Eisele et al., 2018) with permission from Elsevier.