Figure 5.
Altered Localization of VEX1 after ATR RNAi
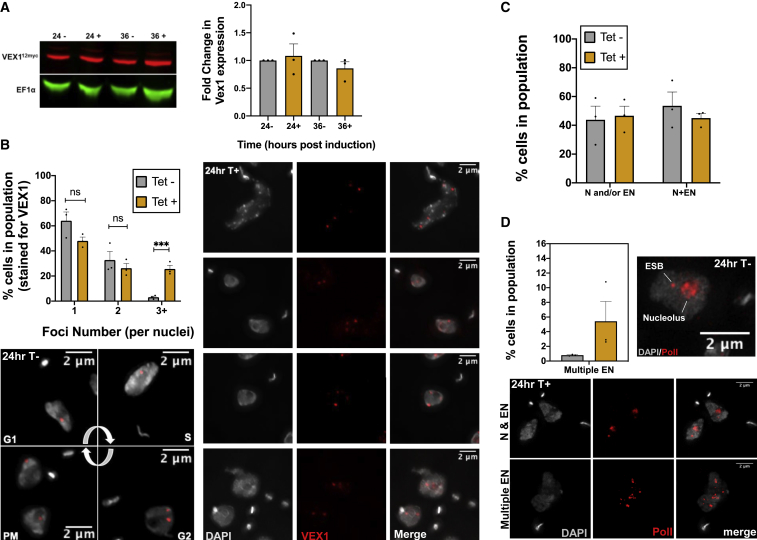
(A) Immunoblot of VEX1−12myc (red) expression after 24 or 36 h of growth with (+) and without (−) RNAi; EF1α (green) serves as a loading control. The graph depicts levels of VEX1−12myc protein after normalization using EF1α (set to 1.0).
(B) Analysis of VEX1−12myc foci number at 24 h of growth with (Tet+, cyan bars) or without (Tet−, gray bars) RNAi. DNA was stained with DAPI and used to determine the number of individual cells harboring 1, 2, or ≥3 (3+) VEX1−12myc foci. Numbers are expressed as a percentage of the total number of cells counted (± SEM). Images show VEX1−12myc localization (red) after 24 h of growth with (T+) and without (T−) RNAi; DAPI-stained DNA is gray (scale bar, 2 μm).
(C) Pol I and ESB after 24 h of growth with (Tet+) and without (Tet−) RNAi. Cells were categorized as having a single subnuclear focus, indicating either nucleolar (N) or extranucleolar staining (EN) staining (N and/or EN), or harboring two clearly distinct foci (N+EN), suggesting both a nucleolus and an ESB; values represent the percentage (± SEM, n = 3) of total cells counted (>100 per experiment).
(D) Analysis of the number of cells harboring >2 extranucleolar foci (multiple EN) per single cell 24 h after RNAi (data plotted as in C). Image on the right is a representative example of Pol I (red) in an uninduced cell, while the images below show representative images of Pol I distribution following RNAi (see also Figure S5; DAPI-stained DNA is gray; scale bar, 2 μm).