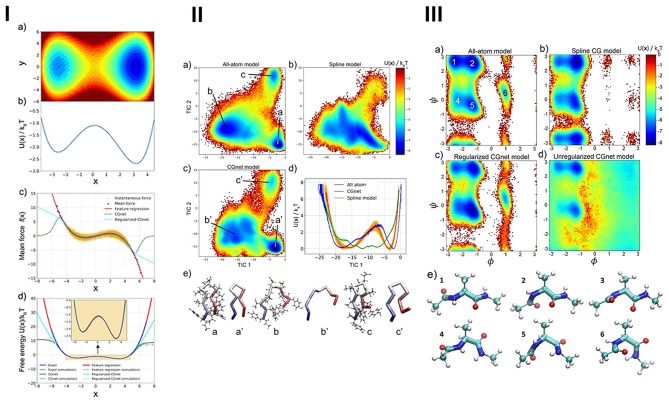
Figure 7.
(I) Machine-learned coarse-graining of dynamics in (a) a two-dimensional potential, showing the (b) exact free-energy along x, comparison of (c) the instantaneous forces and the learned mean forces using feature regression and coarse-grained neural network models with the exact forces, and (d) the potential-of-mean-force along x, predicted by feature regression, and coarse-grained neural network models with the exact free energy. (II) Free-energy profiles and representative structures of alanine dipeptide simulated using all-atom and machine-learned coarse-grained models: (a) free-energy reference as a function of the dihedral angles, obtained from the histograms of all-atom simulations, (b) standard coarse-grained model using a sum of splines of individual internal coordinates, (c) regularized coarse-grained neural network models, (d) unregularized networks, (e) representative structures extracted from the free-energy minima, from atomistic simulation (ball-and-stick representation) and regularized coarse-grained neural network simulation (licorice representation). (III) Free-energy landscape of Chignolin for the different models, obtained from the (a) all-atom simulation, as a function of the first two TICA coordinates, (b) spline model, as a function of the same two coordinates used in the all-atom model, (c) coarse-grained neural network model, as a function of the same two coordinates. (d) Comparison of the one-dimensional free-energy profile as a function of the first TICA coordinate, reflecting the folding/unfolding transition, for the all-atom (blue), spline (green), and coarse-grained neural network models (red). (e) Representative Chignolin conformations in the three minima from (a–c) all-atom simulation and (a′-c′) coarse-grained neural network model. Reprinted with permission from Wang et al. (2019).