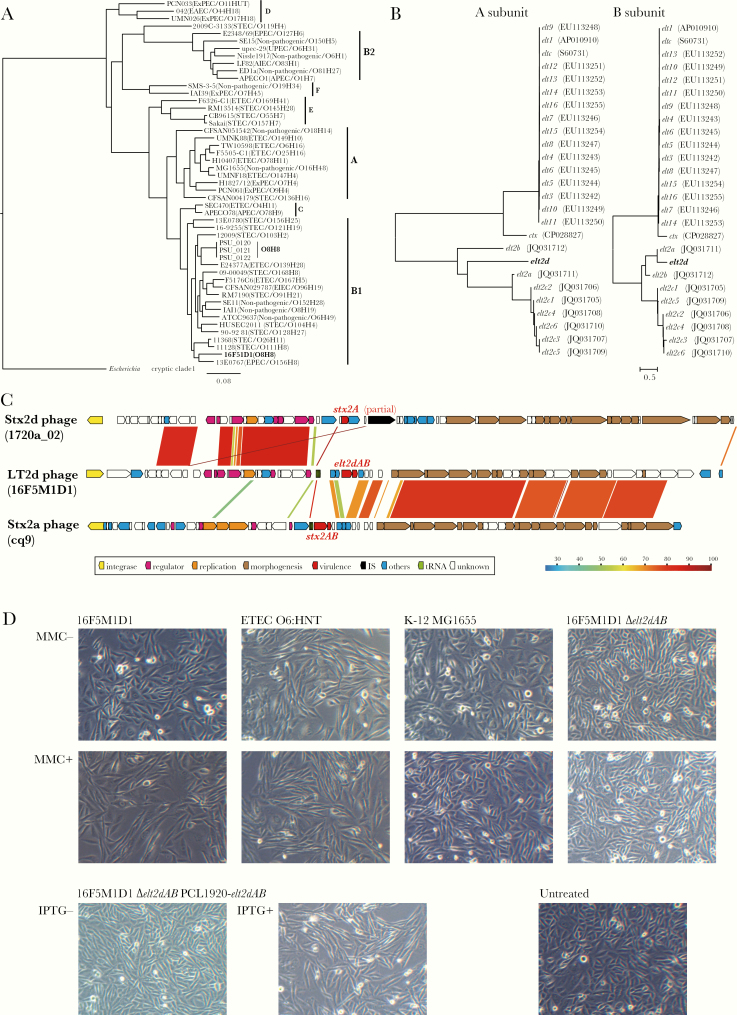
Figure 1.
The phylogenetic analyses, genetic structure of the LT2d-encoding phage, and CHO cell elongation assay. A, A core gene–based maximum likelihood (ML) tree of Escherichia coli O8:H8 strain 16F5M1D1 and an E. coli reference strain set. A cryptic Escherichia clade I strain TW15838 was included as an outgroup. The tree was constructed based on 225 254 SNP sites located on 2569 core genes. Phylogroups, pathotypes, and serotypes are indicated. B, Neighbor-joining (NJ) trees based on the nucleotide sequences of the A and B subunit genes of elt2d and other known elt1 and elt2 variants. The cholera toxin genes (ctx) were also included in this analysis. Accession numbers of each gene are indicated in parentheses. C, The genetic structure of the LT2d-encoding phage is shown. In panel, the genome sequence of the LT2d phage was compared with that of 2 Stx2 phages, to which the LT2d phage genome showed the highest similarity in the early and late regions, respectively. Sequence identities are indicated by different colors. D, The results of the CHO cell elongation assay. CHO cells (2×105 cells/well/500 µL) in a 24-well plate were treated with 100-fold diluted bacterial cell lysates for 48 hours at 37°C and visualized under a light microscope (×100). Cell lysates were prepared by sonicating bacterial cultures incubated with the presence or absence of 500 ng/mL of mitomycin C or 0.1 mM of Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 5 hours at 37°C. Enterotoxigenic E. coli O6:HNT (LT1-positive) and E. coli K-12 MG1655 were used as positive and negative controls, respectively. Untreated: CHO cells untreated with bacterial lysate.