Abstract
Nicotinamide phosphoribosyltransferase (NAMPT) is the rate-limiting enzyme in the NAD+ salvage pathway from nicotinamide. By controlling the biosynthesis of NAD+, NAMPT regulates the activity of NAD+-converting enzymes, such as CD38, poly-ADP-ribose polymerases, and sirtuins (SIRTs). SIRT6 is involved in the regulation of a wide number of metabolic processes. In this study, we investigated the ability of SIRT6 to regulate intracellular NAMPT activity and NAD(P)(H) levels. BxPC-3 cells and MCF-7 cells were engineered to overexpress a catalytically active or a catalytically inactive SIRT6 form or were engineered to silence endogenous SIRT6 expression. In SIRT6-overexpressing cells, NAD(H) levels were up-regulated, as a consequence of NAMPT activation. By immunopurification and incubation with recombinant SIRT6, NAMPT was found to be a direct substrate of SIRT6 deacetylation, with a mechanism that up-regulates NAMPT enzymatic activity. Extracellular NAMPT release was enhanced in SIRT6-silenced cells. Also glucose-6-phosphate dehydrogenase activity and NADPH levels were increased in SIRT6-overexpressing cells. Accordingly, increased SIRT6 levels reduced cancer cell susceptibility to H2O2-induced oxidative stress and to doxorubicin. Our data demonstrate that SIRT6 affects intracellular NAMPT activity, boosts NAD(P)(H) levels, and protects against oxidative stress. The use of SIRT6 inhibitors, together with agents inducing oxidative stress, may represent a promising treatment strategy in cancer.—Sociali, G., Grozio, A., Caffa, I., Schuster, S., Becherini, P., Damonte, P., Sturla, L., Fresia, C., Passalacqua, M., Mazzola, F., Raffaelli, N., Garten, A., Kiess, W., Cea, M., Nencioni, A., Bruzzone, S. SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells.
Keywords: nicotinamide phosphoribosyltransferase, Sirtuin 6, nicotinamide adenine dinucleotide
Nicotinamide phosphoribosyltransferase (NAMPT) is a key enzyme for the synthesis of NAD+. Nicotinamide (NAM) is converted by NAMPT to nicotinamide mononucleotide (NMN), which is then adenylated to NAD+ (1, 2). NAD+ is an essential coenzyme in many reactions, as well as a substrate for NAD+-converting enzymes, such as CD38, poly-ADP-ribose polymerases (PARPs), and sirtuins (SIRTs), a family of NAD+-dependent deacetylases. Thus, by controlling the biosynthesis of the substrate NAD+, NAMPT regulates the activity of these enzymes and the processes they are involved in, including calcium signaling, cellular metabolism, mitochondrial biogenesis, inflammation, stress responses, DNA repair, and circadian rhythm (3, 4). In line with the fundamental requirement of appropriate NAD+ intracellular levels in healthy physiologic conditions, intracellular NAD+ levels have been documented to decrease in metabolic disorders and during ageing in different organs, including pancreas, liver, white adipose tissue, and skeletal muscle (5, 6). In contrast, NAMPT, and consequently NAD+ synthesis, are up-regulated in cancer and in activated immune cells, in which the demand for energy is increased. In line with this notion, NAMPT inhibitors, which strongly decrease NAD+ levels, have been proposed as pharmacological therapeutic interventions in oncologic and immune-mediated disorders (7). NAMPT inhibition with FK866 also affects the NADP(H) pool, although to a different extent, depending on the cell type (8, 9). Notably, in breast cancer cells, the NAMPT-mediated increase in NAD+ levels has been shown to be paralleled by an increase in NADPH through the pentose phosphate pathway, resulting in tumor cell protection from glucose deprivation-induced oxidative stress (10).
In addition to its key role as an intracellular enzyme catalyzing a limiting step in NAD+ biosynthesis, NAMPT is also released by different cell types and extracellular (e)NAMPT has cytokine-like activities (3, 11, 12). Increased levels of eNAMPT have been associated with different metabolic disorders and cancer (3, 13). eNAMPT secretion in adipocytes is increased by SIRT1-mediated deacetylation of NAMPT itself (14). Adipose tissue–specific Nampt-knockout mice exhibit reduced plasma eNAMPT levels and a specific defect in NAD+ biosynthesis in the hypothalamus (14). Overall, intracellular NAMPT activity, intracellular NAD+ levels, and eNAMPT release seem to rely on strict regulation, in that their derangement is associated with pathologic conditions.
SIRT6, another member of the SIRT family, whose activity depends on the availability of intracellular NAD+ (8), is involved in the regulation of a wide variety of processes, including metabolism, cancer, and inflammation (15, 16). Among other SIRT family members, SIRT6 is unique, being endowed with deacetylase activity, but also with a de-fatty acylase activity (17), and also with mono-ADP-ribosyltransferase activity (18, 19).
In cancer, SIRT6 has been reported to play a context-dependent role. In gastrointestinal tumors, SIRT6 acts as a tumor suppressor, through different mechanisms, which include its ability to prevent the Warburg effect and genomic instability. However, other studies indicate that in other types of tumors, including multiple myeloma and skin squamous cell carcinoma, high SIRT6 expression is associated with poor clinical outcome and may act pro-oncogenically (20–22). Dissecting the biologic function of SIRT6 is crucial for a rational exploitation of this target in human disease.
In this study, we sought to find out whether SIRT6 regulates intracellular NAMPT activity, NAD(P)(H) levels, and NAMPT secretion.
MATERIALS AND METHODS
Cell lines
BxPC-3 and MCF-7 cells, engineered to stably overexpress SIRT1 or the catalytically active SIRT6 form [wild-type (WT) SIRT6] or to express a catalytically inactive SIRT6 isoform (SIRT6 H133Y) or transfected with the corresponding empty plasmid (pBABEPURO, pBP), and BxPC-3 cells, transfected with the empty pRETROSUPER (pRS) plasmid or with the pRS SIRT6 sh2 plasmid to down-regulate SIRT6 expression, were cultured as described by Bauer et al. (23).
HEK 293 cells were cultured in DMEM supplemented with 10% fetal calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml) (24). HepG2 cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 IU penicillin, and 100 μg/ml streptomycin. To knockdown SIRT6 in HepG2 cells, we used the Neon Transfection System (100 μl; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s manual. In brief, HepG2 cells were split 1:3 one day before transfection. Cells were transiently transfected with 2 different small interfering RNAs (siRNAs) targeting SIRT6 or with scrambled siRNA control. At 24 h after transfection, the medium was changed for a further 24 or 48 h incubation in serum-free medium at 37°C. All cells were cultured in a humidified 5% CO2 atmosphere at 37°C.
Measurement of NAD(P)(H) levels
BxPC-3 and MCF-7 (pBP, WT SIRT6, SIRT6 H133Y, pRS, and SIRT6 sh2) cells were plated at a density of 2 × 105 cells/well in 12-well plates, cultured for 24 h, harvested, and lysed in 0.1 ml 0.6 M perchloric acid (PCA) (for NAD+ and NADP+) or of 0.1 M NaOH (for NADH and NADPH) at 4°C. Cell extracts were centrifuged for 3 min at 16,000 g, the supernatants were collected, and an aliquot was diluted 20-fold in 100 mM sodium phosphate buffer (pH 8.0), for determination of NAD+ content, as described by Bruzzone et al. (8). NAD(P)(H) values were normalized to protein concentrations, determined by Bradford assay (Bio-Rad Laboratories, Segrate, Italy).
Assay of glucose-6-phosphate dehydrogenase activity
MCF-7 cells (pBP, WT SIRT6, and SIRT6 H133Y) were lysed in ice-cold buffer [25 mM Tris-HCl (pH 7.4), 1 mM EDTA, and protease inhibitors] by brief sonication. The lysates were centrifuged at 10,000 g for 10 min at 4°C. The supernatants (100 μg proteins) were assayed for glucose-6-phosphate dehydrogenase (G6PD) activity at 25°C by measuring the reduction of NADP+ in the reaction buffer [100 mM Tris-HCl (pH 7.4), 0.5 mM EDTA, 10 mM MgCl2, 0.2 mM NADP+, and 0.6 mM glucose 6 phosphate].
Cell viability and cell viability upon oxidative stress
HepG2 cell viability analysis was conducted with the cell proliferation reagent WST-1 (Roche, Basel, Switzerland), according to the manufacturer’s instructions.
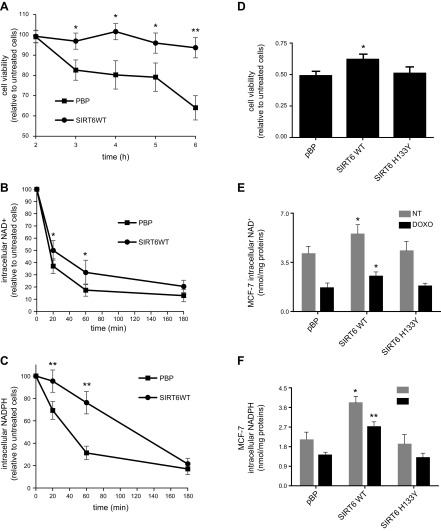
MCF-7 cells (pBP, WT SIRT6, and SIRT6 H133Y) were seeded in 96-well plates (2 × 104 cells/well). The day after, cells were treated (or not) with 2.25 μM doxorubicin for 24 h or with 400 μM H2O2 for 2, 3, 4, 5, and 6 h. Cell viability was evaluated with the sulforhodamine B (SRB) method (25), and NAD+ and NADPH levels were measured as previously described.
NAD+ synthesis assays with cell extracts
Assay of NAD+ synthesis was performed as described by Zoppoli et al. (26). BxPC-3 cells (pBP, WT SIRT6, SIRT6 H133Y, pRS, and SIRT6 sh2) were resuspended in 250 μl NaH2PO4/Na2HPO4 buffer (0.01 M, pH 7.4), frozen at 80°C for 1 h, thawed at room temperature, and sonicated on ice for 10 s at 3 W (W380; Heat-System Ultrasonics, New York, NY, USA), in the presence of protease inhibitors (MilliporeSigma, Burlington, MA, USA). Cell debris was removed by centrifugation at 5000 g for 2 min. Fifty microliters of the lysate was added to a 200-μl reaction mix [50 mM Tris-HCl (pH 7.4) 3 mM ATP, 5 mM MgCl2, 0.5 mM 5-phosphoribosyl 1-pyrophosphate (PRPP), and 2.5 mM NAM] and incubated at 37°C. The reaction was stopped after 2 h by addition of PCA (0.6 M final concentration). NAD+ content was measured by enzymatic cycling assay (27) and the enzymatic activity for NAD+ production expressed as nanomoles per minute per milligram protein. The protein content of each sample was determined with the Bradford assay.
Immunopurification of WT and mutant NAMPT
Plasmids (pCXN2) of FLAG-tagged mouse NAMPT and mutant FLAG-tagged mouse NAMPT (K53R, K79R) were a generous gift from Prof. Shin-Ichiro Imai (Washington University, St. Louis, MO, USA). The full-length cDNAs encoding WT- and K369A-hNAMPT were subcloned by PCR with WT-NAMPT-pET15b plasmid (28) and K369A-NAMPT-pET15b plasmid obtained by site-directed mutagenesis with the QuickChange Lightning Kit (Agilent Technologies, Santa Clara, CA, USA). cDNAs were obtained with the following primers: 5′-AAGCTTATGATCCTGCGGCAGAAGCCGAG-3′ (forward) and 5′-GGATCCCTA ATGATGTGCTGCTTCCAGTTC-3′ (reverse). PCR was performed in 20 µl containing 1× iProof HF reaction buffer, 200 µM dNTP, and 0.5 µM primers and using 0.01 U of iProof HF DNA Polymerase (Bio-Rad Laboratories). The PCR reaction profile was 1 cycle at 98°C for 60 s, 35 cycles at 98°C for 10 s, 60°C for 30 s, and 72°C for 3 min, with a final extension of 5 min at 72°C. The PCR products were purified with a Nucleospin Extraction Kit (Macherey-Nagel, Düren, Germany), digested with HindIII and BamHI restriction enzyme (fast digest enzymes; Thermo Fisher Scientific) and cloned into p3XFLAG-CMV-9 with the Rapid Ligation kit (Roche). These vectors allowed the synthesis of the recombinant WT- and K369A-NAMPT proteins as an N-terminal fusion to the FLAG epitope. The WT- and K369A-NAMPT-p3XFLAG-CMV-9 plasmids were purified with the Endofree Plasmid Maxi Kit (Qiagen, Germantown, MD, USA) and sequenced by TibMolbiol (Genoa, Italy). HEK 293 cells (2 × 107 cells) were transfected, as in Fresia et al. (29), to express different FLAG-tagged NAMPT forms (K53R, K79R, and K369A and the respective WT forms). At 48 h after transfection, the HEK 293 cells were lysed in immunoprecipitation buffer [PBS (pH 7.4), 0.5% NP-40, 1 mM EDTA, 1 mM NaF, 10 mM trichostatin A, 10 mM NAM, 0.5 mM DTT, and protease inhibitor cocktail), and cell extracts were incubated in the presence of monoclonal anti-FLAG-M2–conjugated Dynabeads Protein G (MilliporeSigma) for 2–3 h at 4°C. Immunoprecipitates on anti-FLAG beads were incubated in the absence or presence of 20 μg recombinant SIRT1 (MilliporeSigma) or SIRT6 in the SIRT reaction buffer [20 mM phosphate buffer (pH 7.5) and 2 mM NAD+], for 1 h at 37°C. After incubation, the beads were washed 3 times in PBS and resuspended in the NAMPT reaction buffer [50 mM Tris-HCl (pH 8.5), 100 mM NaCl, 0.25 mM NAM, 10 mM MgSO4, 0.5 mM PRPP, and 2 mM ATP) and incubated (or not, time = 0) for 1.5 h at 37°C. Reactions were stopped by centrifugation and by addition of PCA (0.6 M final concentration) to the supernatants. Supernatants were then neutralized with K2CO3 (250 μM, final concentration), and NMN content was measured by HPLC analysis and by a cycling assay. The pellets, containing the NAMPT-attached Dynabeads, were used for Western blot analysis.
Assay of eNAMPT levels and activity
BxPC-3 and MCF-7 (pRS and SIRT6 sh2) cells were seeded in 24-well plates (8.5 × 104 cells/well). HepG2 cells were seeded in 6-well plates (7 × 105 cells/well) and transfected with siSIRT6 or scrambled siRNA control (as previously described). Cell culture medium was replaced with fresh medium after 24 h and collected after a further 24 h. eNAMPT levels were detected with a commercial kit (AdipoGen Life Sciences, Liestal, Switzerland) or semiquantitatively via Western blot analysis. eNAMPT enzymatic activity in concentrated supernatants of MCF-7 (pRS and SIRT6 sh2) cells was measured as described by Schuster et al. (30).
Transfection of MCF-7 cells with WT and mutant NAMPT-FLAG
MCF-7 (pRS and SIRT6 sh2) cells were transfected with WT or K369A NAMPT-FLAG by using the Nucleofector System (Amaxa, Cologne, Germany). MCF-7 (2 × 106 cells) were transfected with 2 μg of WT-NAMPT-FLAG or K393A-NAMPT-FLAG with Nucleofector Kit V, according to the manufacturer’s instructions (Nucleofector program P-020; Amaxa). After 48 h, cell supernatants were recovered to evaluate eNAMPT levels (as previously described).
Production of recombinant proteins
Recombinant human SIRT6 and NMN adenylyltransferase (NMNAT)-1 were produced as previously described (31, 32).
Determination of Km
Wild-type NAMPT-FLAG was overexpressed, immunopurified, and incubated (or not) with SIRT6, as described above. SIRT6-treated and not treated NAMPT-FLAG Km was determined upon incubation of beads in the NAMPT reaction buffer, containing different NAM concentrations (2, 10, 50, or 250 μM) and plotting the results in a double-reciprocal plot.
Western blot analyses
Cells were washed with cold PBS, collected, and centrifuged at 700 g for 10 min. Cell pellets were lysed in cold lysis buffer [50 mM Tris-HCl, 150 mM NaCl, and 1% NP-40 (pH 7.4)], containing protease and phosphatase inhibitor cocktails (MilliporeSigma). Total protein concentrations were determined by the Bradford method (Bio-Rad Laboratories). Identical amounts of lysate proteins (20 μg/sample) or NAMPT-conjugated Dynabeads were resuspended in SDS sample buffer containing 10% 2-ME, loaded onto SDS 10% polyacrylamide gels, electrophoretically separated and transferred to Immun-Blot PVDF membranes (Bio-Rad Laboratories). Membranes were blocked with 5% nonfat dry milk in PBS for 1 h at room temperature and visualized with the following antibodies: anti-vinculin (a kind gift from E. Turco; Molecular Biotechnology Center, Turin, Italy) anti-acetyl lysine (Cell Signaling Technologies, Danvers, MA, USA), anti-NAMPT (Bethyl Laboratories, Montgomery, TX, USA), anti-FLAG (MilliporeSigma), anti-G6PD (Santa Cruz Biotechnology, Dallas, TX, USA), anti-SIRT6 (MilliporeSigma), and anti-GAPDH (Thermo Fisher Scientific). Secondary antibodies were horseradish peroxidase conjugated (GE Healthcare, Little Chalfont, United Kingdom). Western blots were developed with the ECL-Plus Kit (GE Healthcare), according to the manufacturer’s instructions. Band detection and densitometry were performed with the Chemi-Doc System and the Quantity One software package (Bio-Rad Laboratories).
NMN measurements
NMN synthesis upon incubation of bead suspension (as previously described) was quantified in the neutralized supernatants by analytical phosphate HPLC (25) and by an enzymatic cycling assay, which was set up by modifying an existing method to measure NMNAT activity (33). For the cycling assay, samples (3 μl) were preincubated for 2 h with 27 μl H2O and 15 μl of NMNAT1 reaction buffer [30 mM MgCl2, 6 mM ATP, 320 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/KOH, and 60 ng recombinant NMNAT1] for the conversion of NMN to NAD+. Next, 30 μl cycling reagent used to determine NAD+ content [2% (v/v) ethanol, 100 μg/ml alcohol dehydrogenase, 20 μM resazurin, 5 μg/ml diaphorase, 10 μM flavin mononucleotide, 10 mM NAM, and 100 mM sodium phosphate (pH 8.0)] were added to each sample. A standard curve with known amount of NMN was always run in parallel. The increase in resorufin fluorescence (544 nm excitation, 590 nm emission) was measured every 60 min over a 4 h period, on a fluorescence plate reader (Fluostar Optima; BMG Labtechnologies GmbH, Offenburg, Germany). The cycling assay has a limit of quantification 20 times lower than the HPLC analysis (5 vs. 100 pmol).
NAD+ content in organs
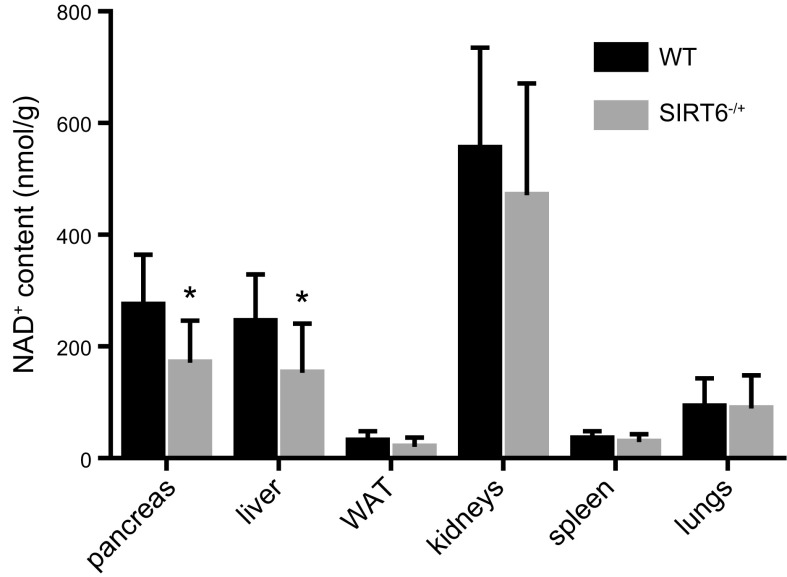
NAD+ levels were evaluated in organs from WT SIRT6−/+ mice. All in vivo experiments were conducted in accordance with the laws and institutional guidelines for animal care, approved by the Institutional Animal Care and Use Committee of the Scientific Institute for Research and Healthcare (IRCCS) University Hospital San Martino–National Institute for Cancer Research (IST). Mice were housed in temperature- and light-controlled conditions (12-h light cycle) with food and water ad libitum. Three- to 9-mo-old 129/Sv Sirt6+/− mice were bred to generate WT and Sirt6−/+mice. Sirt6 knockout mice were not used for this study, because they are known to develop acute metabolic syndrome and die before 4 wk of age (34). Eight WT and 8 Sirt6+/− mice were euthanized when 5-wk-old and liver, pancreas, abdominal adipose tissue, kidneys, spleen, and lungs were collected. Sirt1-deficient (Sirt1−/−) mice (a kind gift from Prof. Greg Zipfel; Washington University, St. Louis, MO, USA) were established as described by Satoh et al. (35). Pancreas, liver, and white adipose tissue were collected from 4 WT and 3 Sirt1−/− mice. Nucleotides were extracted from the organs as described in (36). PCA (0.6 M, final concentration, for NAD+ determination) was added to the minced organs for deproteinization. Samples were neutralized by adding K2CO3, and NAD+ levels were normalized to tissue weight.
Statistical analyses
All parameters were tested by an unpaired Student’s t test. Values of P < 0.05 were considered significant.
RESULTS
SIRT6 expression regulates intracellular NAD+ levels
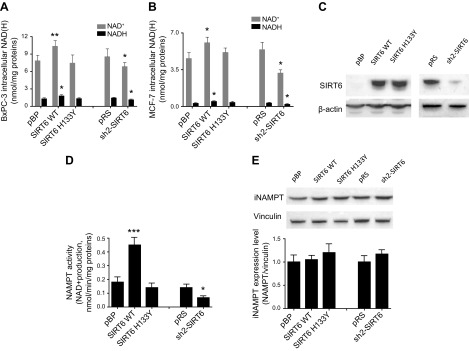
BxPC-3 cells were retrovirally engineered to overexpress a WT, catalytically active form of SIRT6, or to express a catalytically inactive form of the same enzyme (SIRT6 H133Y) (23). Intracellular NAD+ content was found to be significantly, although marginally (by ∼30%), increased in cells overexpressing the WT form of SIRT6, but not in cells expressing the catalytically inactive enzyme (Fig. 1A). In line with this result, the intracellular NAD+ content was decreased when SIRT6 expression was down-regulated by stable RNA interference. Increased NAD+ levels in cells overexpressing SIRT6 were not related to an imbalance in the NAD+/NADH ratio: indeed, the ratio was maintained, and intracellular NADH concentration was increased in SIRT6-overexpressing cells, whereas it was significantly lower in SIRT6-silenced cells. A similar modulation of the NAD+/NADH content was also obtained when SIRT6 was either overexpressed or silenced in MCF-7 cells (Fig. 1B, C).
Figure 1.
A–C) SIRT6 expression modulates the intracellular NAD(H) levels and NAMPT activity. BxPC-3 (A) and MCF-7 (B, C) cells were engineered by retroviral transduction to express the WT form of SIRT6 (SIRT6 WT), or the catalytically inactive SIRT6 mutant (SIRT6H133Y), or were transduced with the empty vector pBP; the same cells were engineered to down-regulate SIRT6 expression (sh2-SIRT6) or were transduced with the respective empty vector pRS. Cells were lysed, and intracellular NAD+ and NADH levels were determined (A, B). The mean ± sd of ≥4 independent determinations. Cells were lysed, and SIRT6 expression level was evaluated by Western blot analysis (representative results are shown) (C). D, E) BxPC-3 cells expressing different levels of SIRT6 were lysed and NAD+ synthesis was evaluated by adding NAM and PRPP as substrates (D), and the iNAMPT expression level was evaluated by Western blot analysis (E). The mean ± sd of results of 3 different analyses, and 1 representative Western blot are shown. *P < 0.05, **P < 0.01, ***P < 0.001 vs. respective controls.
As a comparison, SIRT1 was overexpressed in BxPC-3 cells: NAD+ levels were significantly lower in SIRT1-overexpressing cells, compared to cells transfected with the empty plasmid (from 9.4 ± 2.1 to 5.0 ± 1.7 pmol/mg protein, n = 3, P = 0.04; data not shown). This result is possibly in line with SIRT1 having a much higher NAD+-utilizing deacetylase activity compared to SIRT6 (37, 38), possibly leading to a decrease in the substrate NAD+ in the cells.
SIRT6 regulates NAMPT activity
Because in many cell types NAMPT is the key enzyme responsible for NAD+ biosynthesis, we investigated whether NAD+ production, starting from NAM, would be affected by different levels of SIRT6 expression. In lysates from SIRT6-overexpressing BxPC-3 cells, NAMPT-dependent NAD+ production was increased by 2-fold, as compared to the level measured in lysates from cells transfected with an empty vector (Fig. 1D). Expression of the catalytically inactive SIRT6 did not affect NAM-derived NAD+ synthesis. Accordingly, down-regulation of SIRT6 expression was followed by a decreased ability (by 50%) of BxPC-3 cells to synthesize NAD+ starting from NAM. Two enzymes are responsible for the conversion of NAM to NAD+: NAMPT and NMNAT. The NMNAT-dependent production of NAD+ from lysates of cells expressing different SIRT6 levels was evaluated by adding NMN as the substrate. In this case, no difference was recorded in cells overexpressing SIRT6 or expressing the catalytically inactive SIRT6, as compared to the control cells. Similarly, SIRT6 silencing also did not affect NAD+ synthesis from NMN (data not shown). The level of intracellular NAMPT protein, as measured by Western blot analysis, was not affected in cell lines expressing different SIRT6 levels (Fig. 1E). The results indicate that SIRT6 regulates NAMPT activity, the latter being up-regulated in cells overexpressing SIRT6 and down-regulated in SIRT6-silenced cells, but not its protein level.
NAMPT is a substrate of SIRT6 deacetylase activity
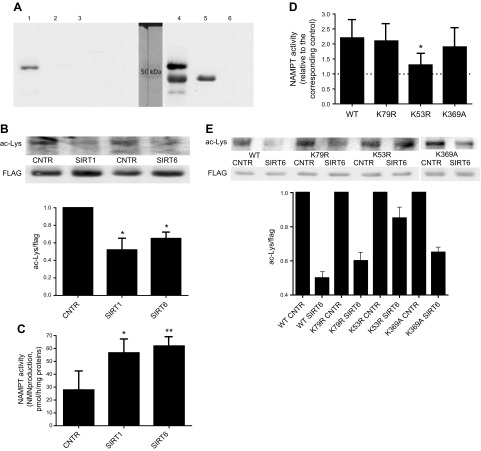
It has been recently demonstrated that NAMPT can be deacetylated by the enzymatic activity of SIRT1 (14). Based on these data, we supposed that SIRT6-mediated regulation of NAMPT activity also relies on its deacetylase activity through direct or indirect mechanisms. To verify the first possibility, a FLAG-tagged NAMPT (FLAG-NAMPT) was expressed in HEK 293 cells (Fig. 2A, lane 1) and purified (lane 4). FLAG-NAMPT was incubated (or not) in the presence of hSIRT6, analyzed by Western blot or further incubated in the presence of its substrates. Western blot analysis revealed that, upon incubation with hSIRT6, the acetylation level of NAMPT was reduced (by ∼50%; Fig. 2B). FLAG-NAMPT, previously incubated or not with hSIRT6, was challenged with its substrates NAM and PRPP, and the production of NMN was quantified by HPLC, as well as by a cycling assay exploiting recombinant NMNAT (which couples the generated NMN to NAD+ production). Both methods showed that the enzymatic activity of FLAG-NAMPT was increased by 2-fold when the protein was deacetylated by hSIRT6 (Fig. 2C). Upon incubation with hSIRT6, NAMPT-FLAG Km for the substrate NAM decreased significantly (from 3.9 ± 0.2 to 1.9 ± 0.1 μM; n = 3), indicating that affinity for NAM is increased when NAMPT is deacetylated by SIRT6. Moreover, FLAG-NAMPT incubation in the presence of hSIRT1 resulted in comparably decreased acetylation level and increased NAMPT activity (Fig. 2B, C). Thus, in the presence of hSIRT6, NAMPT-dependent NMN production was increased compared with that measured with the control NAMPT (not incubated with SIRT6), which is in line with the increased NAD+ content (Fig. 1A, B).
Figure 2.
SIRT6-mediated NAMPT deacetylation increases NAMPT activity. FLAG-NAMPT was expressed in HEK 293 cells and immunoprecipitated with an anti-FLAG antibody. A) Aliquots from the different steps were subjected to Western blot analysis, using the anti-FLAG antibody: a representative result is shown. Lane 1: lysate from FLAG-NAMPT–overexpressing cells; lane 2: supernatant of lysate from FLAG-NAMPT–expressing cells after incubation with magnetic beads; lane 3: lysate from control, not overexpressing, cells; lane 4: eluates from magnetic beads incubated with FLAG-NAMPT–overexpressing cells; lane 5: antibody anti-FLAG; lane 6: supernatant of antibody anti-FLAG after incubation with magnetic beads. B, C) Beads with the immunopurified FLAG-NAMPT were incubated in the absence or the presence of recombinant human SIRT6 or -1, washed, and further incubated in the presence of NAM and PRPP, and NMN synthesis was evaluated by the enzymatic cycling (C). Finally, beads were boiled in loading buffer and subjected to Western blot analysis with an antibody against acetylated lysines or an anti-FLAG antibody, for normalization (B). A representative result and the mean of 4 different determinations are shown. D, E) The WT form, or the K53R, K79R, and K369A mutant forms of FLAG-NAMPT were expressed in HEK 293 cells, immunopurified, incubated (or not, control) with SIRT6, and enzymatic activity (D) and levels of acetylation were determined as previously described (E). *P < 0.05, **P < 0.01 vs. respective controls.
These results demonstrate that NAMPT is a direct substrate of SIRT6 and of SIRT1 activity, with both enzymes regulating NAMPT acetylation level and its enzymatic activity.
Acetylation of NAMPT K53 was demonstrated to reduce NAMPT enzymatic activity (14). To investigate which K residue is important in SIRT6-mediated regulation of NAMPT activity, the WT and mutant forms of NAMPT-FLAG, K53R and K79R, were expressed in HEK cells, immunopurified and incubated (or not) in the presence of hSIRT6. NAMPT activity and its acetylation levels were then analyzed. The enzymatic activity of K53R (normalized to mutant NAMPT protein level, as quantified by Western blot analysis using the anti-FLAG antibody) was increased by ∼1.5-fold compared with wild-type NAMPT, whereas the enzymatic activity of K79R mutants did not differ from that of the WT NAMPT-FLAG (data not shown). When the K79R mutant was incubated in the presence of hSIRT6, an increase in enzymatic activity, similar to that in WT NAMPT, was obtained (Fig. 2D). Instead, when the K53R mutant was incubated with hSIRT6, NAMPT activity did not increase significantly. These results indicate that K53 is a key residue responsible for SIRT6-mediated up-regulation of NAMPT activity. Conversely, K79 did not seem to be involved in SIRT6-mediated regulation of NAMPT enzymatic activity.
Western blot analysis revealed that, upon incubation with hSIRT6, the acetylation level of the K53R NAMPT-FLAG mutant was only slightly reduced (Fig. 2E), suggesting that K53 represents the main (likely not the only) residue that is deacetylated in the WT NAMPT by SIRT6. In contrast, the K79R NAMPT mutant acetylation level was reduced by SIRT6, indicating that K79 is not a main residue substrate for SIRT6 deacetylation.
SIRT6 regulates eNAMPT secretion
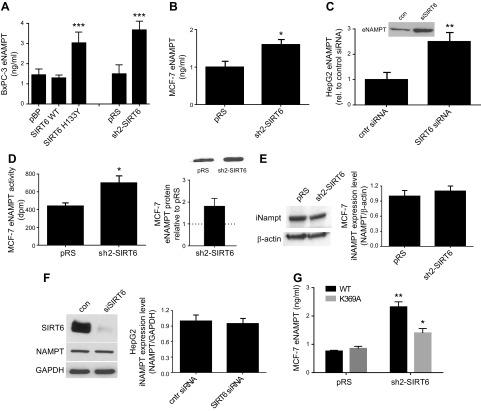
SIRT1-mediated NAMPT deacetylation leads to an increased release of eNAMPT (14). To define whether SIRT6-mediated NAMPT deacetylation also leads to an increase in eNAMPT release, eNAMPT levels were measured in the culture medium of different cell lines, in which SIRT6 expression had been modified. SIRT6 silencing caused a significant increase in eNAMPT level in BxPC-3 (Fig. 3A), in MCF-7 (Fig. 3B), and in HepG2 cell supernatants (Fig. 3C). eNAMPT enzymatic activity was evaluated in supernatants from pRS and sh2-SIRT6 MCF-7 cells (Fig. 3D), and normalized to eNAMPT protein content. However, eNAMPT specific activity was not significantly modified in the 2 conditions. An increase in eNAMPT release was also observed when the catalytically inactive form of SIRT6 was overexpressed in BxPC-3 cells (Fig. 3A). Instead, overexpression of the WT form of SIRT6 did not significantly modify eNAMPT release into the supernatant. Intracellular NAMPT levels were not modified by SIRT6 silencing in MCF-7 cells (Fig. 3E) and in HepG2 cells (Fig. 3F), in line with findings observed in BxPC-3 cells (Fig. 1D). In addition, cell viability was not affected by SIRT6 silencing, indicating that eNAMPT was not released by dying cells (data not shown).
Figure 3.
Down-regulation of SIRT6 expression enhances eNAMPT secretion. A) BxPC-3 cells were engineered by retroviral transduction to express the WT form of SIRT6 (SIRT6 WT), the catalytically inactive SIRT6 mutant (SIRT6H133Y), or the empty vector pBP, or they were engineered to down-regulate SIRT6 expression (sh2-SIRT6) or the respective empty vector pRS. B, D, E) MCF-7 cells were engineered with the vector sh2-SIRT6 (to down-regulate SIRT6 expression) or with the empty vector pRS. C, F) HepG2 cells were transfected with a specific siSIRT6, or with a scramble (scr) siRNA. Cells were seeded in 6-well plates, the supernatants were collected after 48 h and eNAMPT levels were determined by an ELISA kit (A, B) or by Western blot (C, D). eNAMPT enzymatic activity in the supernatant from MCF-7 cells (D). The iNAMPT levels were evaluated by Western blot analysis in McF-7 (E) and in HepG2 lysates (F). A representative result is shown, together with the mean ± sd of 3 independent analyses. G) pRS and sh2-SIRT6 MCF-7 cells were transfected to express the WT or the K369 mutant form of NAMPT-FLAG. eNAMPT release in the supernatant was evaluated by ELISA. All results are means ± sd of results of at ≥3 independent determinations. *P < 0.05, **P < 0.01, ***P < 0.001.
Our data indicate that SIRT6-mediated NAMPT deacetylation resulted in the up-regulation of NAMPT enzymatic activity, similar to the effect exerted by SIRT1. In contrast, with respect to the regulation of eNAMPT release, SIRT1- and SIRT6-mediated regulation seemed to have opposite effects: reduced levels of SIRT6 led to an increased eNAMPT release, whereas SIRT1-mediated NAMPT deacetylation induced an increase in eNAMPT (14). We hypothesized that the acetylation status of different K residues modulates NAMPT activity and eNAMPT release in a different fashion. If a reduced SIRT6 expression causes an increase in eNAMPT release as a consequence of reduced deacetylase activity, one should expect that released eNAMPT possesses at least 1 acetylated K residue. K369 has been reported to be the only acetylated K residue in eNAMPT (14). We reasoned that K369 would be a target for SIRT6-mediated NAMPT deacetylation. Thus, we generated a K369A NAMPT-FLAG mutant. Wild-type and K369A mutant NAMPT-FLAG were overexpressed in pRS and SIRT6-sh2 MCF-7 cells. eNAMPT levels were 3 times higher in supernatant from SIRT6-sh2 MCF-7, compared to the levels in control cells (Fig. 3G). Conversely, SIRT6-mediated regulation of eNAMPT release was lost for K369A NAMPT, suggesting that K369 is a key substrate for SIRT6-mediated regulation of eNAMPT release. To investigate the effect of K369 mutation on NAMPT enzymatic activity, the K369A NAMPT-FLAG was overexpressed in HEK 293 cells and immunopurified. The enzymatic activity of K369A mutant (normalized to the protein level, as quantified by Western blot analysis with the anti-FLAG antibody) did not differ from that of the WT NAMPT-FLAG (data not shown). When the K369A mutant was incubated in the presence of hSIRT6, an increase in enzymatic activity, similar to that obtained in wild-type NAMPT, was observed (Fig. 2D), indicating that K369 is not critical for SIRT6-mediated regulation of NAMPT activity. Western blot analysis revealed that, upon incubation with hSIRT6, the acetylation level of the K369A NAMPT mutant was reduced by SIRT6, apparently to a lower extent compared with the wild-type NAMPT form (Fig. 2E), suggesting that K369 is a substrate for SIRT6 mediated deacetylation. Specifically, K369 deacetylation appeared not to affect NAMPT enzymatic activity (Fig. 2D), but to affect eNAMPT release (Fig. 3G).
SIRT6 expression regulates intracellular NADPH levels and confers resistance to oxidative stress
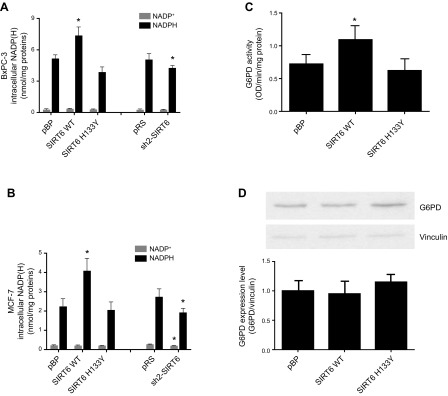
Next, we evaluated whether the level of SIRT6 had an impact on the NADP(H) pools. NADP+ and NADPH levels were evaluated in cells with different levels of SIRT6 expression. In both BxPC-3 and MCF-7 cells, the level of NADPH (Fig. 4A, B) mirrored NAD+ levels (Fig. 1A, B). When SIRT6 was overexpressed, NADPH levels were consistently increased, leading to an increased NADPH:NADP+ ratio in cells with high SIRT6 levels: when SIRT6 was down-regulated, NADPH levels were significantly decreased. NADPH is mainly produced by the pentose phosphate pathway, with G6PD representing the rate-limiting enzyme of this pathway. G6PD enzymatic activity was measured in cell lysates and it proved to be significantly enhanced in cells overexpressing the catalytically active SIRT6 isoform (Fig. 4C). G6PD activity was not affected by SIRT6 silencing (data not shown). G6PD protein levels were not modified by SIRT6 overexpression (Fig. 4D), indicating that higher protein levels of SIRT6 up-regulated G6PD enzymatic activity.
Figure 4.
SIRT6 expression modulates the intracellular NADP(H) levels and G6PD activity (but not its expression). A, B) BxPC-3 (A) and McF-7 (B) cells were engineered by retroviral transduction to express the WT form of SIRT6 (SIRT6 WT), the catalytically inactive SIRT6 mutant (SIRT6H133Y), or the empty vector pBP. The same cells were engineered to down-regulate SIRT6 expression (sh2-SIRT6) or the respective empty vector pRS. Cells were lysed, and intracellular NADP+ and NADPH levels were evaluated. Data are means ± sd of results of 4 independent determinations. C, D) MCF-7 cell lysates were used to evaluate G6PD enzymatic activity (C) or G6PD expression levels (D). The means ± sd from 3 different enzymatic measurements are shown, and a representative Western blot is shown. *P < 0.05.
SIRT6-overexpressing MCF-7 cells proved to be more resistant to H2O2-induced oxidative stress (Fig. 5A), possibly as a consequence of the increased NADPH levels. In addition, SIRT6 overexpression conferred resistance to doxorubicin (Fig. 5D), an anticancer drug, with activity that is, at least in part, mediated by the induction of reactive oxygen species production in cells (39, 40). Intracellular NAD+ and NADPH levels were measured in H2O2 (Fig. 5B, C)- and doxorubicin-treated cells (Fig. 5E, F). In SIRT6-overexpressing cells, the induced reduction in NAD+ levels, as expected for the activation of PARP, and especially in NADPH content, were significantly less prominent. These data are in line with SIRT6 sustaining higher NAD+ and NADPH levels through activation of NAMPT and G6PD and conferring resistance to oxidative stress.
Figure 5.
SIRT6 expression confers resistance to H2O2- and doxorubicin-induced cell death. A–C) MCF-7 cells were seeded in a 96-well plate (A) or in a 12-well plate (B, C) and exposed (or not) to 400 μM H2O2 for the indicated times. Cell were then fixed with trichloroacetic acid and stained with SRB to evaluate cell viability (A), or lysed to measure the intracellular NAD+ (B) and NADPH (C) content. D–F) MCF-7 cells were seeded in a 96-well plate (D), or in a 12-well plate (E, F) and exposed (or not) to 2.25 μM doxorubicin for 24 h. Cell were then fixed with trichloroacetic acid and stained with SRB to evaluate cell viability (D), or lysed to measure the intracellular NAD+ (E) and NADPH (F) content. Cell viability was determined in quadruple. Values represent the mean ± sd of 4 independent determinations. *P < 0.05, **P < 0.01.
SIRT6 expression levels regulate NAD+ levels in different organs
To investigate whether SIRT6 boosts NAD+ levels in physiologic conditions, and not only in cancer cells, NAD+ content was determined in different organs collected from WT or SIRT6+/− mice. NAD+ levels were significantly decreased in liver and pancreas from SIRT6+/− mice compared with the same organs from WT animals (Fig. 6). On the other hand, no difference between Sirt6+/− and WT mice in NAD+ content was observed in white adipose tissue, kidneys, spleen, and lungs, suggesting that SIRT6-mediated regulation of NAD+ content is organ specific. Notably, in liver and white adipose tissue, but not in pancreas, from SIRT1−/− mice, the NAD+ content was significantly higher (1.27- and 1.34-fold in liver and white adipose tissue, respectively) than in the same organs from WT littermates (data not shown). These data are in line SIRT1 being endowed with a much higher NAD+-utilizing deacetylase activity SIRT6 (37, 38) and with data obtained in SIRT1-overexpressing BxPC3 cells. Moreover, in adipose tissue SIRT1 stimulates eNAMPT release (14), which may also account for the observed intracellular NAD+ decrease.
Figure 6.
Intracellular NAD+ content is reduced in liver and pancreas from SIRT6+/− mice. The indicated organs were harvested from WT or SIRT6+/− mice. Organs were minced in PCA, and NAD+ levels were evaluated. Results are expressed as nanomoles NAD+ normalized to tissue weight (g), and represent means ± sd (n = 8 WT and n = 8 SIRT6+/− mice). *P < 0.05.
Thus, SIRT6 activity appears to boost NAD+, possibly counteracting the NAD+ decrease observed during aging and metabolic disorders.
DISCUSSION
The link between NAD+ levels and SIRT activity has been widely explored, especially SIRT1, with activity that is fueled by NAMPT and which regulates key cellular processes, including cellular energy metabolism, aging, and circadian rhythm (41, 42). SIRT1 and NAMPT have been described to regulate each other’s expression and activity in a positive-feedback manner, with SIRT1 inducing the expression of NAMPT and with NAMPT recycling NAM to provide the SIRT substrate NAD+. In the circadian clock, BMAL1 regulates the circadian expression of NAMPT, with SIRT1 inducing the overexpression of NAMPT and thereby contributing to the circadian synthesis of its own substrate NAD+ (43). A positive-feedback loop between SIRT1 and NAMPT has also been reported to exist in human cancer cells with deregulated expression of c-MYC (44). However, to the best of our knowledge, whether and how other SIRTs regulate NAMPT-mediated NAD+ biosynthesis remains unknown. In our study, SIRT6 activity did not affect NAMPT protein levels (Figs. 1E and 3E, F). Instead, SIRT6 overexpression boosted the synthesis of its own substrate by enhancing NAMPT activity and consequently increasing the NAD pool, especially NADPH (Figs. 1A, B and 4A, B). The mechanism for such an effect likely consists in NAMPT’s being a direct substrate of SIRT6 deacetylase activity (Fig. 2). SIRT1 was also reported to deacetylate NAMPT, with the evidence coming from cell systems in which the 2 proteins were overexpressed (14). In line with that study, we unequivocally demonstrated that NAMPT is a substrate for both SIRT6 and SIRT1, leading to the significant up-regulation of NAMPT activity (Fig. 2C). K53 was demonstrated to be the substrate of SIRT1 deacetylation (14), and, according to our data, the same lysine seems to represent the key residue for SIRT6-mediated NAMPT activation (Fig. 2D). Indeed, K53 is peculiarly located at the “cleft” containing the catalytic sites in the active dimeric form (14). A possible concern is whether SIRT6 and NAMPT are present in the same cell compartment: SIRT6 is mainly reported as a nuclear SIRT (15), although a small fraction of this SIRT was also detected in the cytoplasm and in the endoplasmic reticulum (18, 45, 46). On the other hand, NAMPT localizes to both cytosol and nucleus (47–49). Thus, these studies show that an encounter between the two proteins and the regulation of NAMPT activity by SIRT6 could well occur both in the nucleus and in the cytoplasm.
SIRT6-mediated up-regulation of NAD+ levels (although marginal, by ∼30%) represents a positive loop whereby SIRT6 would increase its own substrate, but at the same time, it would allow the fueling of the other NAD+-consuming enzymes, including other SIRT family members, CD38 and the PARPs. The up-regulation of G6PD activity in SIRT6 overexpressing cells may be related to a direct G6PD protein modification by SIRT6 through one of its enzymes: deacetylase, deacylase, or ADP-ribosyltransferase. Alternatively, the increased G6PD activity may be caused by augmented SIRT2 activity in cells with increased NAD+ levels, because SIRT2 has been reported to activate G6PD in acute myeloid leukemia cells (50). SIRT2 inhibition suppressed G6PD activity, reducing proliferation of leukemia cells, indicating that SIRT2 may represent a promising target for therapeutic applications in leukemia. SIRT6 inhibition has also been proposed as a treatment for leukemia, in view of the disruption of SIRT6-mediated DNA-repair mechanisms, in combination with DNA damaging agents (51).
In this study we suggested that SIRT6 can prevent oxidative stress damage in cells through up-regulation of NAMPT activity and increase in NADPH, as a consequence of G6PD activation. This result is in line with a study reporting that, in breast cancer cells, NAMPT activity counteracts glucose deprivation-induced oxidative stress by inducing an increase in NADPH (10). Thus, our observations indicate that SIRT6 is a key upstream player regulating NAMPT activity in these events. In addition to this mechanism, in human mesenchymal cells, it has been demonstrated that SIRT6 can prevent oxidative stress by means of transactivating NRF2-regulated antioxidant genes, including heme oxygenase 1 (52).
SIRT6 was demonstrated to activate PARP1, with PARP1 being directly ADP-ribosylated by SIRT6 (19). Thus in SIRT6-overexpressing cells, PARP1 activity could be further enhanced in case of oxidative stress. Nevertheless, in these conditions, the levels of NAD+ could be kept significantly higher, at least in the first phases of oxidative stress (Fig. 5B), likely as a balanced result of SIRT6-mediated activation of both NAMPT and PARP1.
It should be underlined that different approaches to boost NAD+ levels are being investigated as promising means to promote healthy aging and life span (6), given that NAD+ levels decline with age and dysmetabolism/dysfunctions. Three main strategies to boost NAD+ levels are currently under investigation: supplementation of NAD+ precursors (with NR and NMN being promising for the lack of side effects); activation of NAD+ synthesis (with NAMPT and NMNAT representing the most studied enzymes in this respect); and inhibition of NAD+ consumption (with CD38 and PARP1 inhibition having a major impact on NAD+ levels). The SIRT6-mediated regulation of NAMPT activity documented by our study is well in line with the search for tools and mechanisms to sustain NAD+ biosynthesis and positively impact aging and prevention of different aging-related disorders. Along this line, NAD+ content regulation by SIRT6 appears to occur specifically in liver and pancreas (Fig. 6). Thus, the SIRT6-mediated boost of NAD+ levels may contribute to the maintenance of optimal liver functions. This positive effect seems to be organ specific, because SIRT6 expression level did not affect NAD+ content in white adipose tissue, lungs, spleen, and kidneys. Further investigations are needed to define the mechanism(s) underlying this specificity of effect by SIRT6. Indeed, in addition to the up-regulation of NAMPT activation, other changes in NAD+-synthesizing/-utilizing enzymes may be relevant and ultimately result in altered NAD+ levels at different SIRT6 expression levels and degrees of activation.
Our finding that SIRT6 regulates eNAMPT release apparently in a way opposite SIRT1 is intriguing. Although the physiologic significance remains to be identified, it is tempting to speculate that the activity of SIRT1 and -6 in regulating eNAMPT release may be finely coordinated to achieve opposite results in different physiologic and pathologic conditions. That reducing SIRT6 expression determines an increased eNAMPT release implies that an increased acetylation state on 1 or some residues may occur in the secreted protein. K369 is the only acetylated lysine residue in extracellular NAMPT (14), and our study provides evidence that K369 may be relevant for the SIRT6-mediated regulation of eNAMPT release (Fig. 3G). It is also possible that additional mechanisms, related to the other SIRT6 enzymatic activities (i.e., deacylase and ADP-ribosyl transferase), play a role in the regulation of eNAMPT release. Indeed, further studies are necessary to investigate whether NAMPT is acylated and may represent a substrate for SIRT6-mediated deacylation, as occur in the TNF-α release, regulated by SIRT6 through a post-translational mechanism (46). Similarly, the possibility that NAMPT is a substrate for SIRT6 ADP-ribosyl transferase activity, with consequent modulation of its enzymatic activity and release, cannot be ruled out.
As previously mentioned, in several tumors, SIRT6 acts as a tumor suppressor, and different mechanisms have been proposed to underlie this property (16, 20). The ability of SIRT6 to reduce the release of eNAMPT, which has been reported to exert protumorigenic effects (11, 13, 53, 54), could represent a new key mechanism, whereby SIRT6 counters tumorigenesis. In contrast, other neoplasms may take advantage of an enhanced iNAMPT activity via SIRT6 to increase their NAD+ availability. Thus, in principle, both reduced and increased SIRT6 activity could be exploitable by cancer cells, depending on the biologic context.
That a catalytically inactive form of SIRT6 promoted eNAMPT release, as obtained with SIRT6 silencing, may be in line with evidence suggesting that overexpression of the catalytically dead SIRT6 mutant probably exerts a dominant negative effect, thus causing, at least in some respects, effects that are the opposite of those induced by the overexpression of WT SIRT6 (55,56). In our hands, the mutated, catalytically inactive SIRT6 and wild-type SIRT6 only caused opposite effects when it came to regulating eNAMPT release, but not, for instance, when we monitored intracellular NAMPT activity and NAD(P) pools, suggesting that the mechanism by which SIRT6 effects eNAMPT release differs from the one regulating NAMPT activity.
In summary, we demonstrated that SIRT6 regulates intracellular NAMPT activity and NAD(P)(H) levels. Our data shed light on new, potential mechanisms of SIRT6-mediated cancer cell survival and oxidative stress resistance, but they also pinpoint an additional mechanism whereby SIRT6 could have a positive impact on aging.
ACKNOWLEDGMENTS
The authors thank Prof. Antonio De Flora (University of Genoa) for helpful discussions; Prof. Giuseppe Orsomando (Polytechnic University of Marche) for providing the plasmid pTrcHisA-NMNAT1; Prof. Shin-Ichiro Imai (Washington University, St. Louis, MO, USA) for providing the plasmids of FLAG-tagged NAMPT (WT and mutant forms K53R and K79); Prof. Greg Zipfel (Washington University) for providing the SIRT1 KO mice; and Prof. Raul Mostoslavsky (Harvard Medical School, Boston, MA, USA) for providing Sirt6+/− mice. This work was supported by the Italian Association for Cancer Research (AIRC) Grants IG AIRC 17736 (to A.N.) and GR-2011-02347192 (to A.N. and A. Grozio); Athero-B-Cell Grant FP7 GA 602114 (to A.N.); and Pancreas Grant GA 56986 (to A.N. and S.B.); the Fondazione Umberto Veronesi (to A.N.); U.S. Army, Department of Defense, Breast Cancer Research Program Grant BC161452P1 (to A.N.); 5×1000 Funds from the Hospital Policlinico San Martino, Scientific Institute for Research and Healthcare (IRCCS) for Oncology (to M.C. and A.N.); and the University of Genova (to S.B.). I.C. and P.D. are supported by Fondazione Umberto Veronesi. G.S. is recipient of a fellowship for young investigators granted by Ghislieri College (Pavia, Italy). The authors declare no conflicts of interest.
Glossary
- eNAMPT
extracellular nicotinamide phosphoribosyltransferase
- FLAG-NAMPT
FLAG-tagged nicotinamide phosphoribosyltransferase
- G6PD
glucose-6-phosphate dehydrogenase
- NAM
nicotinamide
- NAMPT
nicotinamide phosphoribosyltransferase
- NMN
nicotinamide mononucleotide
- NMNAT
NMN adenylyltransferase
- PARP
poly-ADP-ribose polymerase
- PCA
perchloric acid
- PRPP
5-phosphoribosyl-1(α)-pyrophosphate
- siRNA, small interfering RNA
SIRT, sirtuin
- SRB
sulforhodamine B
- WT
wild type
AUTHOR CONTRIBUTIONS
S. Bruzzone designed the research; G. Sociali, A. Grozio and P. Becherini performed experiments and analyzed the data; I. Caffa, S. Schuster, P. Damonte, L. Sturla, C. Fresia, F. Mazzola, and M. Passalacqua performed experiments; S. Bruzzone, N. Raffaelli, A. Garten, W. Kiess, M. Cea, and A. Nencioni analyzed the data; and S. Bruzzone drafted the manuscript.
REFERENCES
- 1.Magni G., Amici A., Emanuelli M., Raffaelli N., Ruggieri S. (1999) Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73, 135–182, xi [DOI] [PubMed] [Google Scholar]
- 2.Dölle C., Skoge R. H., Vanlinden M. R., Ziegler M. (2013) NAD biosynthesis in humans: enzymes, metabolites and therapeutic aspects. Curr. Top. Med. Chem. 13, 2907–2917 [DOI] [PubMed] [Google Scholar]
- 3.Garten A., Schuster S., Penke M., Gorski T., de Giorgis T., Kiess W. (2015) Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 11, 535–546 [DOI] [PubMed] [Google Scholar]
- 4.Nikiforov A., Kulikova V., Ziegler M. (2015) The human NAD metabolome: functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 50, 284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshino J., Mills K. F., Yoon M. J., Imai S. (2011) Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajman L., Chwalek K., Sinclair D. A. (2018) Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metab. 27, 529–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montecucco F., Cea M., Bauer I., Soncini D., Caffa I., Lasigliè D., Nahimana A., Uccelli A., Bruzzone S., Nencioni A. (2013) Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors as therapeutics: rationales, controversies, clinical experience. Curr. Drug Targets 14, 637–643 [DOI] [PubMed] [Google Scholar]
- 8.Bruzzone S., Fruscione F., Morando S., Ferrando T., Poggi A., Garuti A., D’Urso A., Selmo M., Benvenuto F., Cea M., Zoppoli G., Moran E., Soncini D., Ballestrero A., Sordat B., Patrone F., Mostoslavsky R., Uccelli A., Nencioni A. (2009) Catastrophic NAD+ depletion in activated T lymphocytes through Nampt inhibition reduces demyelination and disability in EAE. PLoS One 4, e7897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song T. Y., Yeh S. L., Hu M. L., Chen M. Y., Yang N. C. (2015) A Nampt inhibitor FK866 mimics vitamin B3 deficiency by causing senescence of human fibroblastic Hs68 cells via attenuation of NAD(+)-SIRT1 signaling. Biogerontology 16, 789–800 [DOI] [PubMed] [Google Scholar]
- 10.Hong S. M., Park C. W., Kim S. W., Nam Y. J., Yu J. H., Shin J. H., Yun C. H., Im S. H., Kim K. T., Sung Y. C., Choi K. Y. (2016) NAMPT suppresses glucose deprivation-induced oxidative stress by increasing NADPH levels in breast cancer. Oncogene 35, 3544–3554 [DOI] [PubMed] [Google Scholar]
- 11.Soncini D., Caffa I., Zoppoli G., Cea M., Cagnetta A., Passalacqua M., Mastracci L., Boero S., Montecucco F., Sociali G., Lasigliè D., Damonte P., Grozio A., Mannino E., Poggi A., D’Agostino V. G., Monacelli F., Provenzani A., Odetti P., Ballestrero A., Bruzzone S., Nencioni A. (2014) Nicotinamide phosphoribosyltransferase promotes epithelial-to-mesenchymal transition as a soluble factor independent of its enzymatic activity. J. Biol. Chem. 289, 34189–34204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamporlini F., Ruggieri S., Mazzola F., Amici A., Orsomando G., Raffaelli N. (2014) Novel assay for simultaneous measurement of pyridine mononucleotides synthesizing activities allows dissection of the NAD(+) biosynthetic machinery in mammalian cells. FEBS J. 281, 5104–5119 [DOI] [PubMed] [Google Scholar]
- 13.Carbone F., Liberale L., Bonaventura A., Vecchiè A., Casula M., Cea M., Monacelli F., Caffa I., Bruzzone S., Montecucco F., Nencioni A. (2017) Regulation and function of extracellular nicotinamide phosphoribosyltransferase/visfatin. Compr. Physiol. 7, 603–621 [DOI] [PubMed] [Google Scholar]
- 14.Yoon M. J., Yoshida M., Johnson S., Takikawa A., Usui I., Tobe K., Nakagawa T., Yoshino J., Imai S. (2015) SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab. 21, 706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kugel S., Mostoslavsky R. (2014) Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 39, 72–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lerrer B., Gertler A. A., Cohen H. Y. (2016) The complex role of SIRT6 in carcinogenesis. Carcinogenesis 37, 108–118 [DOI] [PubMed] [Google Scholar]
- 17.Feldman J. L., Baeza J., Denu J. M. (2013) Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liszt G., Ford E., Kurtev M., Guarente L. (2005) Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320 [DOI] [PubMed] [Google Scholar]
- 19.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. (2011) SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tasselli L., Zheng W., Chua K. F. (2017) SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 28, 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cea M., Cagnetta A., Adamia S., Acharya C., Tai Y. T., Fulciniti M., Ohguchi H., Munshi A., Acharya P., Bhasin M. K., Zhong L., Carrasco R., Monacelli F., Ballestrero A., Richardson P., Gobbi M., Lemoli R. M., Munshi N., Hideshima T., Nencioni A., Chauhan D., Anderson K. C. (2016) Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood 127, 1138–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ming M., Han W., Zhao B., Sundaresan N. R., Deng C. X., Gupta M. P., He Y. Y. (2014) SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 74, 5925–5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer I., Grozio A., Lasigliè D., Basile G., Sturla L., Magnone M., Sociali G., Soncini D., Caffa I., Poggi A., Zoppoli G., Cea M., Feldmann G., Mostoslavsky R., Ballestrero A., Patrone F., Bruzzone S., Nencioni A. (2012) The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J. Biol. Chem. 287, 40924–40937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruzzone S., Basile G., Chothi M. P., Nobbio L., Usai C., Jacchetti E., Schenone A., Guse A. H., Di Virgilio F., De Flora A., Zocchi E. (2010) Diadenosine homodinucleotide products of ADP-ribosyl cyclases behave as modulators of the purinergic receptor P2X7. J. Biol. Chem. 285, 21165–21174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grozio A., Sociali G., Sturla L., Caffa I., Soncini D., Salis A., Raffaelli N., De Flora A., Nencioni A., Bruzzone S. (2013) CD73 protein as a source of extracellular precursors for sustained NAD+ biosynthesis in FK866-treated tumor cells. J. Biol. Chem. 288, 25938–25949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoppoli G., Cea M., Soncini D., Fruscione F., Rudner J., Moran E., Caffa I., Bedognetti D., Motta G., Ghio R., Ferrando F., Ballestrero A., Parodi S., Belka C., Patrone F., Bruzzone S., Nencioni A. (2010) Potent synergistic interaction between the Nampt inhibitor APO866 and the apoptosis activator TRAIL in human leukemia cells. Exp. Hematol. 38, 979–988 [DOI] [PubMed] [Google Scholar]
- 27.Bruzzone S., De Flora A., Usai C., Graeff R., Lee H. C. (2003) Cyclic ADP-ribose is a second messenger in the lipopolysaccharide-stimulated proliferation of human peripheral blood mononuclear cells. Biochem. J. 375, 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buonvicino D., Mazzola F., Zamporlini F., Resta F., Ranieri G., Camaioni E., Muzzi M., Zecchi R., Pieraccini G., Dölle C., Calamante M., Bartolucci G., Ziegler M., Stecca B., Raffaelli N., Chiarugi A. (2018) Identification of the nicotinamide salvage pathway as a new toxification route for antimetabolites. Cell Chem. Biol. 25, 471–482.e7 [DOI] [PubMed] [Google Scholar]
- 29.Fresia C., Vigliarolo T., Guida L., Booz V., Bruzzone S., Sturla L., Di Bona M., Pesce M., Usai C., De Flora A., Zocchi E. (2016) G-protein coupling and nuclear translocation of the human abscisic acid receptor LANCL2. Sci. Rep. 6, 26658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster S., Penke M., Gorski T., Petzold-Quinque S., Damm G., Gebhardt R., Kiess W., Garten A. (2014) Resveratrol differentially regulates NAMPT and SIRT1 in hepatocarcinoma cells and primary human hepatocytes. PLoS One 9, e91045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sociali G., Galeno L., Parenti M. D., Grozio A., Bauer I., Passalacqua M., Boero S., Donadini A., Millo E., Bellotti M., Sturla L., Damonte P., Puddu A., Ferroni C., Varchi G., Franceschi C., Ballestrero A., Poggi A., Bruzzone S., Nencioni A., Del Rio A. (2015) Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur. J. Med. Chem. 102, 530–539 [DOI] [PubMed] [Google Scholar]
- 32.Sorci L., Cimadamore F., Scotti S., Petrelli R., Cappellacci L., Franchetti P., Orsomando G., Magni G. (2007) Initial-rate kinetics of human NMN-adenylyltransferases: substrate and metal ion specificity, inhibition by products and multisubstrate analogues, and isozyme contributions to NAD+ biosynthesis. Biochemistry 46, 4912–4922 [DOI] [PubMed] [Google Scholar]
- 33.Revollo J. R., Grimm A. A., Imai S. (2004) The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279, 50754–50763 [DOI] [PubMed] [Google Scholar]
- 34.Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M. M., Mills K. D., Patel P., Hsu J. T., Hong A. L., Ford E., Cheng H. L., Kennedy C., Nunez N., Bronson R., Frendewey D., Auerbach W., Valenzuela D., Karow M., Hottiger M. O., Hursting S., Barrett J. C., Guarente L., Mulligan R., Demple B., Yancopoulos G. D., Alt F. W. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 [DOI] [PubMed] [Google Scholar]
- 35.Satoh A., Brace C. S., Ben-Josef G., West T., Wozniak D. F., Holtzman D. M., Herzog E. D., Imai S. (2010) SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 30, 10220–10232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montecucco F., Bauer I., Braunersreuther V., Bruzzone S., Akhmedov A., Lüscher T. F., Speer T., Poggi A., Mannino E., Pelli G., Galan K., Bertolotto M., Lenglet S., Garuti A., Montessuit C., Lerch R., Pellieux C., Vuilleumier N., Dallegri F., Mage J., Sebastian C., Mostoslavsky R., Gayet-Ageron A., Patrone F., Mach F., Nencioni A. (2013) Inhibition of nicotinamide phosphoribosyltransferase reduces neutrophil-mediated injury in myocardial infarction. Antioxid. Redox Signal. 18, 630–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan P. W., Feldman J. L., Devries M. K., Dong A., Edwards A. M., Denu J. M. (2011) Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldman J. L., Dittenhafer-Reed K. E., Kudo N., Thelen J. N., Ito A., Yoshida M., Denu J. M. (2015) Kinetic and structural basis for acyl-group selectivity and NAD(+) dependence in sirtuin-catalyzed deacylation. Biochemistry 54, 3037–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang W. P., Chau S. P., Kong S. K., Fung K. P., Kwok T. T. (2003) Reactive oxygen species mediate doxorubicin induced p53-independent apoptosis. Life Sci. 73, 2047–2058 [DOI] [PubMed] [Google Scholar]
- 40.Luanpitpong S., Chanvorachote P., Nimmannit U., Leonard S. S., Stehlik C., Wang L., Rojanasakul Y. (2012) Mitochondrial superoxide mediates doxorubicin-induced keratinocyte apoptosis through oxidative modification of ERK and Bcl-2 ubiquitination. Biochem. Pharmacol. 83, 1643–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai S., Guarente L. (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., Bass J. (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menssen A., Hydbring P., Kapelle K., Vervoorts J., Diebold J., Lüscher B., Larsson L. G., Hermeking H. (2012) The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc. Natl. Acad. Sci. USA 109, E187–E196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao N. N., Ren J. H., Tang H., Ran L. K., Zhou H. Z., Liu B., Huang A. L., Chen J. (2017) Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 485, 713–719 [DOI] [PubMed] [Google Scholar]
- 46.Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R., Hang H. C., Hao Q., Lin H. (2013) SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rongvaux A., Shea R. J., Mulks M. H., Gigot D., Urbain J., Leo O., Andris F. (2002) Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32, 3225–3234 [DOI] [PubMed] [Google Scholar]
- 48.Kitani T., Okuno S., Fujisawa H. (2003) Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett. 544, 74–78 [DOI] [PubMed] [Google Scholar]
- 49.Zhang T., Berrocal J. G., Frizzell K. M., Gamble M. J., DuMond M. E., Krishnakumar R., Yang T., Sauve A. A., Kraus W. L. (2009) Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J. Biol. Chem. 284, 20408–20417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu S. N., Wang T. S., Li X., Wang Y. P. (2016) SIRT2 activates G6PD to enhance NADPH production and promote leukaemia cell proliferation. Sci. Rep. 6, 32734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cagnetta A., Soncini D., Orecchioni S., Talarico G., Minetto P., Guolo F., Retali V., Colombo N., Carminati E., Clavio M., Miglino M., Bergamaschi M., Nahimana A., Duchosal M., Todoerti K., Neri A., Passalacqua M., Bruzzone S., Nencioni A., Bertolini F., Gobbi M., Lemoli R. M., Cea M. (2018) Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells’ vulnerability to DNA-damaging agents. Haematologica 103, 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W., Li Y., Yang J., Hao Y., Yuan T., Yuan G., Wang H., Ju Z., Mao Z., Li J., Qu J., Tang F., Liu G. H. (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 26, 190–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grolla A. A., Torretta S., Gnemmi I., Amoruso A., Orsomando G., Gatti M., Caldarelli A., Lim D., Penengo L., Brunelleschi S., Genazzani A. A., Travelli C. (2015) Nicotinamide phosphoribosyltransferase (NAMPT/PBEF/visfatin) is a tumoural cytokine released from melanoma. Pigment Cell Melanoma Res. 28, 718–729 [DOI] [PubMed] [Google Scholar]
- 54.Audrito V., Serra S., Brusa D., Mazzola F., Arruga F., Vaisitti T., Coscia M., Maffei R., Rossi D., Wang T., Inghirami G., Rizzi M., Gaidano G., Garcia J. G., Wolberger C., Raffaelli N., Deaglio S. (2015) Extracellular nicotinamide phosphoribosyltransferase (NAMPT) promotes M2 macrophage polarization in chronic lymphocytic leukemia. Blood 125, 111–123 [DOI] [PubMed] [Google Scholar]
- 55.Grimley R., Polyakova O., Vamathevan J., McKenary J., Hayes B., Patel C., Smith J., Bridges A., Fosberry A., Bhardwaja A., Mouzon B., Chung C. W., Barrett N., Richmond N., Modha S., Solari R. (2012) Over expression of wild type or a catalytically dead mutant of sirtuin 6 does not influence NFκB responses. PLoS One 7, e39847; erratum, doi.org/10.1371/annotation/9c1a5765-6471-401f-bc74-a916c8ead46f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang N., Li Z., Mu W., Li L., Liang Y., Lu M., Wang Z., Qiu Y., Wang Z. (2016) Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle 15, 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]