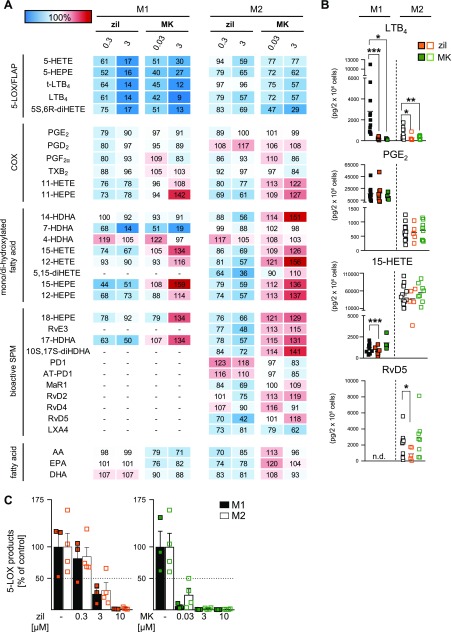
Figure 3.
Effects of the 5-LOX inhibitor zileuton (zil) and the FLAP inhibitor MK886 (MK) on LM biosynthesis in M1 and M2. A) Human M1 and M2 (2 × 106 cells/ml) were preincubated with 0.3 or 3 µM zileuton, 0.03 or 3 µM MK886, or vehicle (0.1% DMSO) for 15 min at 37 °C and then incubated with E. coli (O6:K2:H1; ratio 1:50) for another 180 min. Formed LMs were extracted by SPE and analyzed by UPLC-MS-MS, and means are shown in a heat map as percentage of vehicle-treated cells (= 100% control, white). For zil, n = 8 in M1 (n = 4 for 14-HDHA, 7-HDHA, 12-HETE, and 17-HDHA; n = 6 for 15-HEPE and 12-HEPE; n = 7 for 4-HDHA, 18-HEPE, and DHA) and n = 5 in M2 (n = 4 for LXA4). For MK, n = 4 in M1 and n = 8 in M2 (n = 6 for RvD2; n = 7 for MaR1; n = 4 for LXA4). B) Effects of 3 µM zileuton (zil, orange) and 3 µM MK886 (MK, green) on LTB4, PGE2, 15-HETE, and RvD5 biosynthesis in M1 and M2, shown as pg/2 × 106 cells. Data were log-transformed for paired Student’s t test. N.d., not detectable. Zil vs. vehicle in M1: LTB4, ***P = 0.000006; PGE2, P = 0.274; 15-HETE, ***P = 0.00016, and in M2: LTB4, *P = 0.0475; PGE2, P = 0.896; 15-HETE, P = 0.0797; RvD5, *P = 0.0125. MK vs. vehicle in M1: LTB4, *P = 0.0234; PGE2, P = 0.329; 15-HETE, P = 0.1324, and in M2: LTB4, **P = 0.0083; PGE2, P = 0.706; 15-HETE, P = 0.0719; RvD5, P = 0.2800. C) M1 and M2 (2 × 106 cells/ml) were preincubated with zileuton, MK886, or vehicle (0.1% DMSO) for 15 min at 37 °C and then incubated with A23187 (2.5 µM) for another 10 min. Formed 5-LOX products (LTB4, its isomers, 5-HETE, 5S,6R-diHETE, and 5-HEPE) were extracted by SPE and analyzed by UPLC-MS-MS. Data are given as means + sem, n = 4 independent experiments, shown as percentage of vehicle control (= 100%).