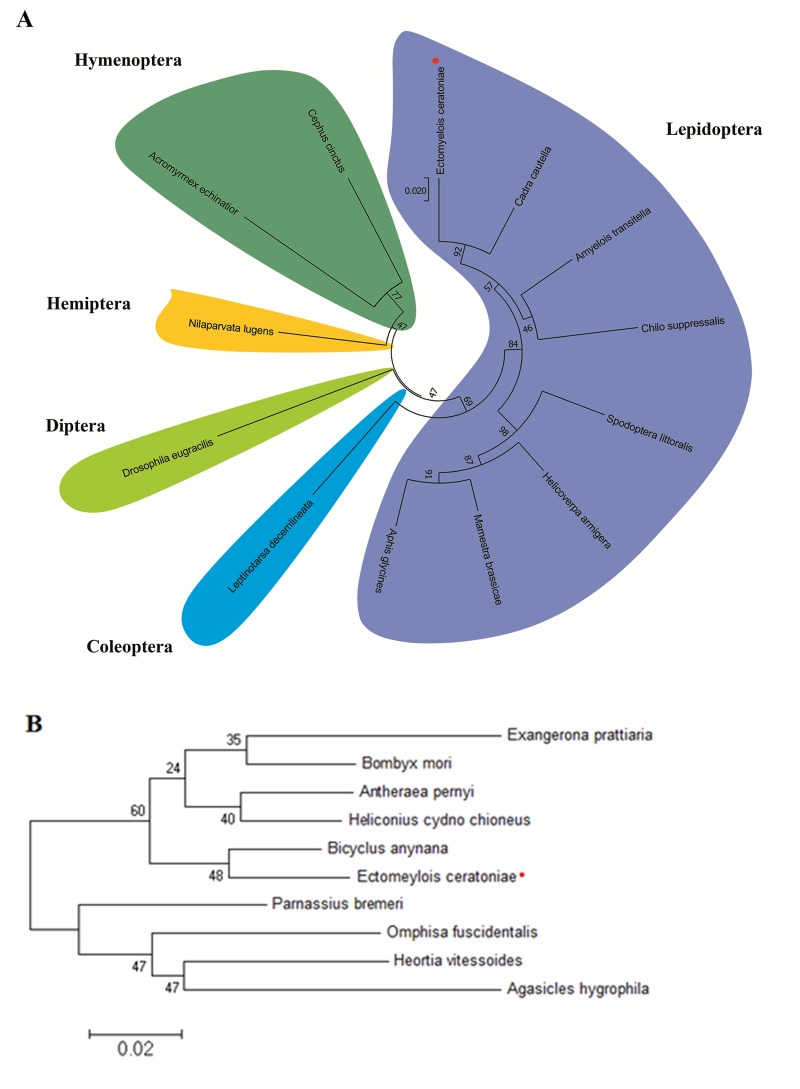
Fig 1. Phylogenetic relationship of sequences of HSP70 and HSP90 of carob moth and other insects.
(A) An unrooted neighbor-joining tree of HSP70 cDNA sequences was constructed with bootstrap support based on 10,000 replications shown at the relevant branch points. Cadra cautella (MF067297.1), Amyelois transitella (XM_013331933.1), Mamestra brassicae (AB251896.1), Helicoverpa armigera (XM_021342205.1), Chilo suppressalis (GU726137.1), Spodoptera littoralis (KC787696.1), Leptinotarsa decemlineata (KC544268.1), Drosophila eugracilis (XM_017212762.1), Nilaparvata lugens (KX976471.1), Acromyrmex echinatior (XM_011062182.1), Cephus cinctus (XM_015747637.2). (B) Neighbor-joining tree of predicted protein sequences of HSP90 from the carob moth and other lepidopteran insects. Heortia vitessoides (AWX67685.1), Exangerona prattiaria (ADM26739.1), Antheraea pernyi (APX61060.1), Parnassius bremeri (APP93215.1), Agasicles hygrophila (ALP48323.1), Bombyx mori (AEB39782.1), Omphisa fuscidentalis (ABP93404.1), Bicyclus anynana (AFM73651.1), Heliconius cydno chioneus (AFQ40718.1).