Abstract
Cortical circuit activity is shaped by the parvalbumin (PV) and somatostatin (SST) interneurons that inhibit principal excitatory (EXC) neurons and the vasoactive intestinal peptide (VIP) interneurons that suppress activation of other interneurons. To understand the molecular-genetic basis of functional specialization and identify potential drug targets specific to each neuron subtype, we performed a genome wide assessment of both gene expression and splicing across EXC, PV, SST and VIP neurons from male and female mouse brains. These results reveal numerous examples where neuron subtype-specific gene expression, as well as splice-isoform usage, can explain functional differences between neuron subtypes, including in presynaptic plasticity, postsynaptic receptor function, and synaptic connectivity specification. We provide a searchable web resource for exploring differential mRNA expression and splice form usage between excitatory, PV, SST, and VIP neurons (http://research-pub.gene.com/NeuronSubtypeTranscriptomes). This resource, combining a unique new dataset and novel application of analysis methods to multiple relevant datasets, identifies numerous potential drug targets for manipulating circuit function, reveals neuron subtype-specific roles for disease-linked genes, and is useful for understanding gene expression changes observed in human patient brains.
SIGNIFICANCE STATEMENT Understanding the basis of functional specialization of neuron subtypes and identifying drug targets for manipulating circuit function requires comprehensive information on cell-type-specific transcriptional profiles. We sorted excitatory neurons and key inhibitory neuron subtypes from mouse brains and assessed differential mRNA expression. We used a genome-wide analysis which not only examined differential gene expression levels but could also detect differences in splice isoform usage. This analysis reveals numerous examples of neuron subtype-specific isoform usage with functional importance, identifies potential drug targets, and provides insight into the neuron subtypes involved in psychiatric disease. We also apply our analysis to two other relevant datasets for comparison, and provide a searchable website for convenient access to the resource.
Keywords: interneuron, psychiatric, PV, splicing, SST, VIP
Introduction
Circuit activity in the cortex and hippocampus is shaped by distinct contributions from specific interneuron subpopulations (Isaacson and Scanziani, 2011). Recently, cell-type-specific manipulation using optogenetic approaches has provided insight into the circuit roles of three classes of interneurons: those that express parvalbumin (PV), somatostatin (SST), or vasoactive intestinal peptide (VIP; Kepecs and Fishell, 2014). While PV, SST, and VIP neurons can be further subdivided, and other interneuron subtypes also exist, the majority of interneurons fall into these three broad, non-overlapping classes (Rudy et al., 2011; Zeisel et al., 2015; Tasic et al., 2016). PV interneurons provide feedforward inhibition to pyramidal neurons and control the temporal fidelity and synaptic integration of excitatory inputs (Pouille and Scanziani, 2001; Gabernet et al., 2005), and are critical for the generation of gamma oscillations (Bartos et al., 2007; Cardin et al., 2009; Sohal et al., 2009). SST interneurons are spontaneously active and provide powerful regulation of local neuronal activity through dense connections to nearby pyramidal neurons (Urban-Ciecko and Barth, 2016). VIP interneurons exert their influence on network function via a dis-inhibitory circuit such that when driven by local, long-range, or neuromodulatory inputs, VIP neurons inhibit SST interneurons (and to a lesser extent PV interneurons), resulting in reduced inhibition of pyramidal neurons (Lee et al., 2013; Pi et al., 2013; Karnani et al., 2016).
Consistent with important roles in circuit regulation, dysfunction of cortical interneurons has been implicated in various psychiatric and cognitive disorders. For example, data from mouse models and human postmortem tissue point to PV neuron dysfunction in schizophrenia (SCZ; Lewis et al., 2012; Gonzalez-Burgos et al., 2015), and PV and SST interneuron dysfunction are seen in Alzheimer's disease (AD) mouse models (Ramos et al., 2006; Verret et al., 2012; Palop and Mucke, 2016; Schmid et al., 2016). Interneuron dysfunction is also implicated in autism spectrum disorder (ASD) and bipolar disorder (BD; Marín, 2012; Takano, 2015). Consistent with the roles of PV and SST interneurons in inhibition of pyramidal neurons, reduced function of these interneurons occurs in animal models of epilepsy (Tai et al., 2014; Huusko et al., 2015). Conversely, reducing activation of VIP interneurons (which normally disinhibit excitatory neurons) can oppose seizure induction in animal models (Khoshkhoo et al., 2017). Given the roles in diverse nervous system diseases, it is of great interest to identify therapeutic targets that could selectively modulate these interneuron subtypes widely across the cortex.
Recent studies profiling neurons from specific brain regions using single-cell RNA-seq and sorted population sequencing strategies have improved our understanding of neuron subtype-specific gene expression (Zeisel et al., 2015; Mardinly et al., 2016; Tasic et al., 2016; Paul et al., 2017; Hrvatin et al., 2018). However, despite the importance of alternative splicing in the nervous system (Raj and Blencowe, 2015), the current understanding of interneuron subtype-specific spliceform usage is superficial. Large-scale, genome-wide analysis of splicing across neuron subtypes has been limited by (1) the low throughput of long-read technologies; (2) few datasets containing both excitatory neurons and inhibitory neuron subtypes with sufficient coverage at splice junctions; and (3) a lack of methods that can use biological replicates and harness splice junction data, rather than relying upon inference from exonic coverage.
To identify druggable targets with interneuron subtype-specific expression across the cortex, and to comprehensively assess alternative splicing across neuron subtypes, we used FACS to isolate cortical neurons (neocortex + archicortex) from transgenic mice expressing fluorescent proteins in excitatory neurons (EXC), or PV, SST, or VIP interneurons, and obtained RNA-seq data for each cell type. In these experiments, we used a robust number of biological replicates for each neuron subtype, and used Thy1-GFP mice to sort a population of excitatory neurons that avoids the glial contamination inherent in Emx-cre sorting strategies. We analyzed these data to identify differential gene expression and used graph analysis of splice junction reads to assess differential splicing with more power and specificity than can be obtained by inference from exonic reads. We provide this genome-wide analysis of neuron subtype transcriptomes in a searchable database as an online resource for the community, and include new analysis of relevant published datasets for comparison. This resource and our analysis provide extensive insight into transcriptomic differences correlating with the unique functional properties of interneurons, highlight numerous genes that might represent targets for selective modulation of interneuron and circuit function, and implicate specific neuron subtypes in psychiatric diseases.
Materials and Methods
Mice.
All protocols involving animals were approved by Genentech's Institutional Animal Care and Use Committee, in accordance with guidelines that adhere to and exceed state and national ethical regulations for animal care and use in research. Excitatory neurons were sorted from four male and two female Thy1-GFP-M mice, which exhibit sparse expression of EGFP in excitatory neurons (RRID:IMSR_JAX:007788; Feng et al., 2000). Interneurons were sorted using tdTomato Cre reporter mice (RRID:IMSR_JAX:007914; Madisen et al., 2010) crossed to PV-Cre (RRID:IMSR_JAX:008069; Hippenmeyer et al., 2005; 3 males, 3 females), or Sst-IRES-Cre (RRID:IMSR_JAX:013044; 4 males, 2 females), or Vip-IRES-Cre knock-in mice (RRID:IMSR_JAX:01908; Taniguchi et al., 2011; 2 males, 4 females). For optogenetic experiments, PV-Cre mice were crossed to R26-CAG-LSL-2XChETA-tdTomato mice (RRID:IMSR_JAX:017455). All experiments were performed using 3-month-old mice (mixed male and female). All protocols were approved by the Genentech Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Isolation of neuron subtypes.
Isolation of pure populations of neurons from adult mouse cortex using fluorescence activated cell sorting (FACS) was performed following published protocols (Saxena et al., 2012) with the following modifications: after enzymatic digestion with papain and trituration to obtain a homogenous cell suspension, cells were washed with 5 ml of Hibernate A-LF twice at 1800 × g and resuspended in 5 ml of Hibernate A with 2.5 μl of DAPI (1 μg/ml stock) added for FACS sorting to exclude dead cells. Neurons were sorted as DAPI(−):GFP(+) or DAPI(−):tdTomato(+) (depending on the transgenic reporter line) using a BD FACSAria cell sorter that was setup using a 100 μm nozzle at 20 psi. Sorted cells (collected live) in Hibernate A medium were spun at 2000 × g at 4°C for 5 min and cell pellets were processed for RNA extraction using a Qiagen RNeasy Plus Micro kit. RNA quality was checked for a subset of samples using a fragment analyzer (RQN = 5.1 ± 0.5, mean ± SEM, n = 10) and 500 ng of RNA was submitted for sequencing. The collected cell numbers for each replicate averaged ∼50,000 for PV+ cells, ∼30,000 for SST+ cells, ∼45,000 for VIP+ cells, and ∼50,000 for Thy1-GFP cells.
RNA-sequencing data.
Total RNA was extracted from the sorted neurons, cDNA was generated from up to 25 ng of total RNA using Nugen's RNA-Seq method for low-input RNA samples, Ovation RNA-Seq System V2 (NuGEN). Per the manufacturer's instructions, total RNA was neither depleted of rRNA nor polyA-selected. Generated cDNA was sheared to 150–200 bp size using LE220 focused ultra-sonicator (Covaris). Following cDNA shearing, the size of samples was determined using Bioanalyzer DNA 1000 Kit (Agilent). In addition, sheared cDNA samples were quantified by Qubit dsDNA BR Assay (Life Technologies). One microgram of sheared cDNA was taken into further processing, starting at end repair step, using Illumina's TruSeq RNA Sample Preparation Kit v2. From this each 50 bp, single-end RNA-seq library was produced with 8 amplification cycles. We collected six separate FACS samples, with one mouse per sample, for each of the four neuron subtypes, for a total of 24 RNA-seq libraries. Each library was sequenced on an Illumina HiSeq or Rapid HiSeq machine, and resulted in 17–43 million reads per sample. RNA-sequencing quality control metrics, including analysis of 5′/3′ coverage bias are provided in Fig. 1-1. The 5′/3′ bias was consistent across samples in this study. Samples were sequenced in batches with RNA-seq libraries from other tissue sources. We observed small-batch effects in an EXC sample that was sequenced with four VIP samples, and with two VIP samples sequenced with five SST samples. Overall the samples clearly clustered by neuron subtype and we did not exclude any samples. We include the batch group identifier (POOLID) along with each sample in our dataset.
Read processing and mapping.
The fastq sequence files for all RNA-seq samples, including the previously published samples, were then filtered for read quality (keeping reads where at least 70% of the cycles had Phred scores ≥23), and ribosomal RNA contamination. The remaining reads were aligned to the mouse reference genome (GRCm38) using the GSNAP alignment tool (Wu and Nacu, 2010). Alignments were produced using the following GSNAP parameters: “-M 2 -n 10 -B 2 -i 1 -N 1 -w 200000 -E 1 -pairmax-rna = 200000 -clip-overlap”. These steps, and the downstream processing of the resulting alignments to obtain read counts and RPKMs per gene (over exons of RefSeq gene models), are implemented in the Bioconductor package, HTSeqGenie v4.10.0. Only uniquely mapped reads were used for further analysis.
Differential expression analysis.
Initial differential expression analysis was performed with voom+limma (Law et al., 2014; Ritchie et al., 2015). To obtain genes that were enriched in a given neuron subtype, that subtype was compared, pairwise, to each of the other three neuron subtypes. If the gene was significantly differentially enriched (log2(fold-change) ≥ 1, adjusted p ≤ 0.05) in the neuron subtype-of-interest, in all three pairwise comparisons, it was called enriched in the subtype-of-interest.
Splicing analysis.
Splicing events were predicted and quantified by SGSeq (Goldstein et al., 2016). SGSeq yields “splicing events” (or just “events”), which are composed of multiple (but usually just 2) variants. For example, a classical cassette alternative exon would be considered a single event, and the inclusion and skipping isoforms considered the “splice variants” (or just “variants”). The variants in turn are composed of “splice graph features” (or just “features”). A single count for each variant in each sample was determined as previously described in methods for SGSeq (Goldstein et al., 2016). A variant frequency (variantFreq) statistic was calculated by dividing the counts for a particular splice variant by the sum of all variant counts for the particular splice event. For a classical cassette alternative exon event, the two possible variants would be (1) including or (2) skipping the middle exon. Thus, the variantFreq statistic for the inclusion variant would be the number of read counts supporting the inclusion of the cassette exon divided by the sum of read counts supporting the inclusion or skipping of the cassette exon. The variantFreq statistic, which ranges from 0 to 1 can be interpreted as the variant inclusion or exclusion, and is used for visualization purposes throughout the manuscript and website figures. In the case where there are no reads within a sample that cover any variant at a given splice event, the variantFreq cannot be calculated, and the data point for that sample is omitted from any figure for the splice event. Example sequencing coverage at splice junctions is provided in Fig. 4-1 using the splice events featured in Figure 4 as examples.
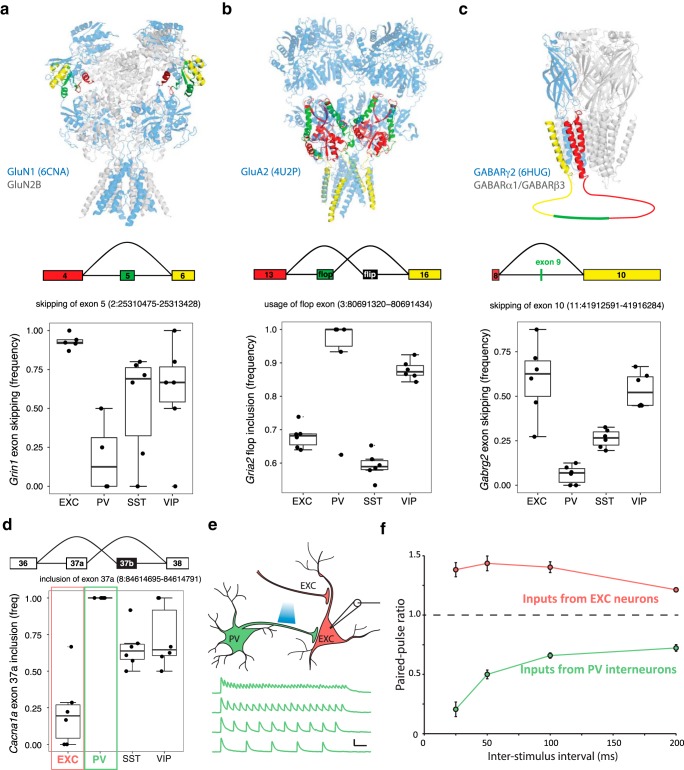
Figure 4.
Examples of subtype-specific exon usage in ligand- and voltage-gated ion channels. a–c, Neuron subtype-specific exon usage in NMDA, AMPA, and GABAA receptors. Homologous regions of protein structures corresponding to the approximate location of alternatively spliced exons and adjacent exons are color coded to match the splicing schematics below. Genomic locations of the splicing events are indicated. Boxplots depict the frequency of the indicated splicing events. Boxes show median and interquartile ranges, and whiskers show full data without outliers. a, Grin1 and its protein product GluN1 (6CNA, rat GluN1-GluN2B heteromer depicted). b, Gria2 and its protein product GluA2 (4U2P, rat GluA2 homomer depicted). The simplified canonical flip/flop alternative splicing in Gria2 is depicted and quantified here. c, Gabrg2 and its protein product GABARγ2 (6HUG, human α1β3γ2L GABAAR depicted). d–f, Neuron subtype-specific alternative splicing in Cacna1a (Cav2.1). d, The frequency of Cacna1 exon 37a inclusion is quantified in the boxplot. Red and green boxes highlight the exclusion of exon 37a (exon 37b inclusion) in EXC neurons and the inclusion of exon 37a in PV neurons, respectively. e, Short-term plasticity of synaptic transmission from PV neurons was measured by recording IPSCs from CA1 pyramidal neurons in response to optogenetic stimulation of PV neurons. Example current traces in response to stimulation at 25, 50, 100, and 200 Hz are shown. Scale bars: 100 pA, 1 s. f, Quantification of the paired pulse ratio between the first and second stimuli at each interval is shown in green (n = 7 recordings with 25, 50, 100, and 200 ms intervals). As predicted from exon 37 isoform usage, pronounced short-term plasticity of PV neuron output was observed, whereas control experiments measuring excitatory neuron output demonstrated short-term facilitation (n = 14 recordings with all intervals). All data in f are shown as mean ± SEM.
Statistical analysis of differential splicing.
To identify differential splicing events, the read counts per sample for each splice variant (as described in the previous section) were analyzed using a variation on the DEXSeq package (Anders et al., 2012). Only variants with at least five counts in at least three samples (of any neuron subtype) were analyzed. Then, any events for which only a single variant remained were considered to be effectively constitutive and that remaining variant was discarded. Following this, DEXSeq was run on the remaining data using the SGSeq event as the DEXSeq “gene” and the SGSeq variant as the DEXSeq “exon”. The model used was sample+exon+neuron subtype: exon. After running DEXSeq, to limit the number of tests, the first variant of each event (generally a “skipping” variant) was discarded (e.g., removing the redundancy of reciprocal variants of the same event). Results were then further filtered to require |log2(fold-change)| > 0.32 (adjusted p < 0.05). This analysis was conducted first for each neuron subtype versus the other three subtypes together (e.g., EXC vs PV, SST, VIP), and then as pairs (e.g., EXC, PV vs SST, VIP).
Statistical analysis of neurexin splicing.
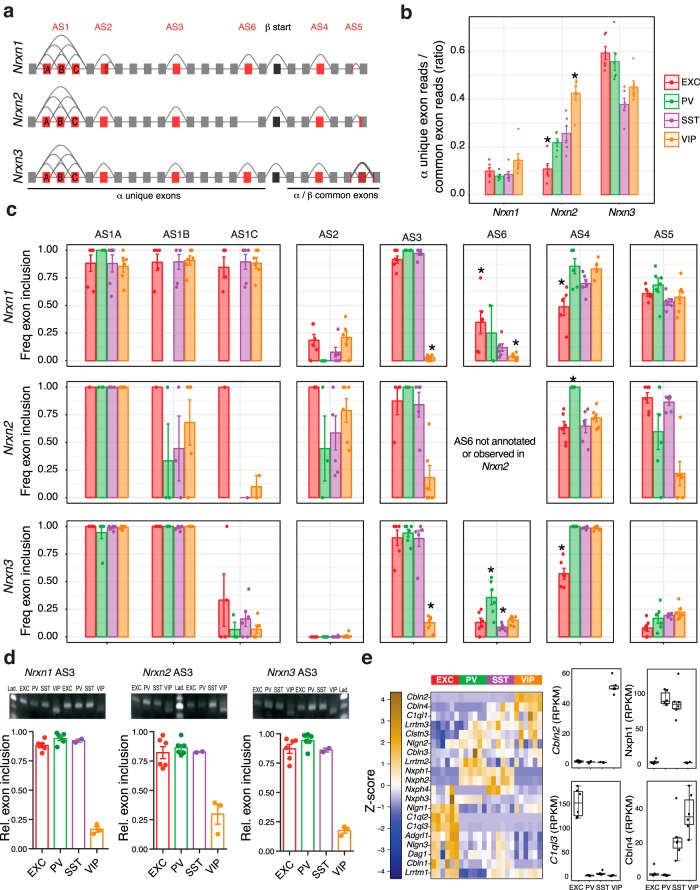
As a proxy for α–β isoform ratios of the neurexin genes, we quantified the ratio of read counts within exons found only the α isoforms to the read counts within exons common to both α and β isoforms. Read counts were summed per sample, across the exons unique to α isoforms for each neurexin gene. Summed read counts per sample were then calculated for the exons in common to both α and β isoforms for each neurexin gene. Ratios were calculated by dividing the sum of α exon counts by the sum of common exon counts (see Fig. 4b). We tested for statistically significant differences in the ratios between neuron subtypes within a neurexin gene by performing unpaired t tests of the ratios for a given neuron subtype (n = 6) versus the other three subtypes (n = 18). For the comprehensive splicing analysis of the neurexin genes (see Fig. 4c), because of the complexity of splice events within neurexins, we performed specific splice junction analysis using known annotations for the six canonical alternative splicing events (AS1–AS6). Providing these splice annotations to SGSeq, variant frequencies were calculated as previously described in Splicing analysis. When variantFreq could not be calculated (due to insufficient read coverage at the event in a sample), that sample was omitted from the plot and from downstream statistical analysis. To test for significance in the splicing variant frequencies unpaired t tests were performed on the exon inclusion frequencies (variantFreq) within a given neurexin gene and splice event, between one neuron subtype (n ≤ 6) and the other three neuron subtypes (n ≤ 18).
Nrxn splicing analysis by RT-PCR.
RNA from sorted cells was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad). Standard PCR amplification (30 cycles) was performed using TaqDNA polymerase and dNTP pack from Roche. One microliter of cDNA was added per 10 μl reaction. The PCR products were run on 2% agarose gels (E-Gel, ThermoFisher Scientific). The following primers were used for PCR: Nrxn1 forward: TCTGCAGACACCCTTCGATT, Nrxn1 reverse: GGCTTGTTGATCATCCACGG, Nrxn2 forward: ACGCTTCATGTCTCAGAGGG, Nrxn2 reverse: ACCGTGTGCCATTCATTGTC, Nrxn3 forward: TTCATGTCACAGCGAGCCTA, Nrxn3 reverse: TGCATACAGGGTCTCAGGTC. The PCR products were run on 2% agarose gels (E-Gel, ThermoFisher Scientific) and gels were imaged using Gel Doc EZ imager (Bio-Rad). PCR band intensities were quantified with ImageJ software after background subtraction.
Short-term plasticity measurements.
Brain slices were prepared as previously described (Hackos et al., 2016) using 3-month-old PV-Cre × R26-CAG-LSL-2XChETA-tdTomato mice. For measurements of PV neuron output whole-cell recordings were made from morphologically identified hippocampal pyramidal neurons and PV neurons were activated using 8 ms flashes of blue light delivered from a Xenon arc lamp (Lambda DG-4, Sutter Instrument) at various stimulation intervals. The timing of light flashes was controlled by custom software developed using MATLAB (MathWorks). Postsynaptic IPSCs were recorded by voltage-clamping at 0 mV in the presence of 10 μm NBQX and 50 μm AP5 to block AMPAR and NMDAR currents, respectively. For measurements of EXC neuron outputs, EPSPs were measured from CA1 pyramidal neurons in response to electrical stimulation of Schaffer collateral inputs using standard extracellular field recording measurements in stratum radiatum. Recording solution compositions were as follows (in mm): ACSF: 127 NaCl, 2.5 KCl, 1.3 MgCl2, 2.5 CaCl2, 1.25 NaH2PO4, 25 NaHCO3, 25 glucose; intracellular solution: 115 Cs-methanesulfonate, 10 CsCl, 10 HEPES, 10 BAPTA-Cs, 2.5 MgCl2, 5 QX314-Cl, 4 Na2ATP, 0.4 Na2GTP, Na2-phosphocreatine.
Comparison of neuronal gene expression to human patient expression profiles.
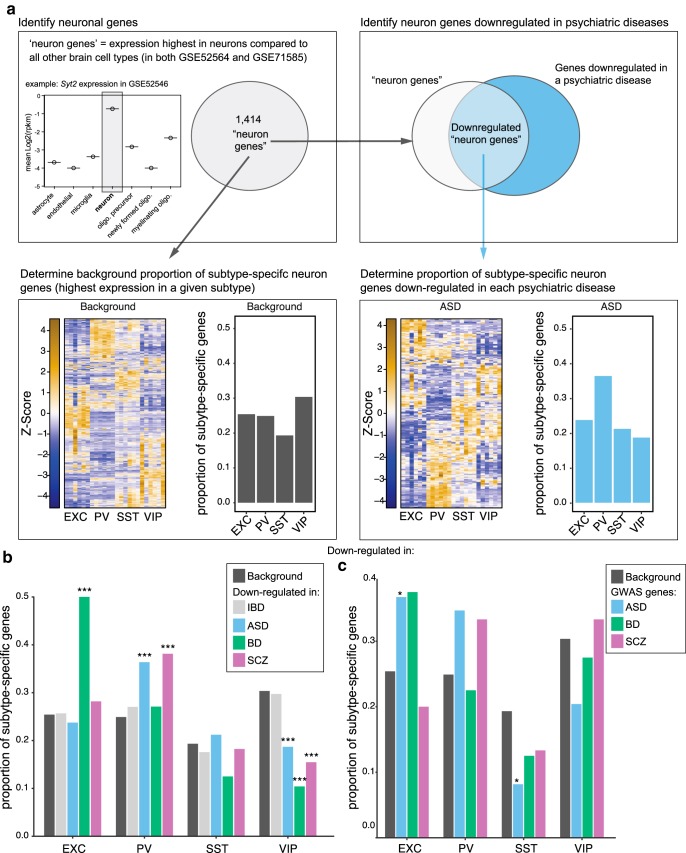
The list of significantly downregulated genes (FDR < 0.05) from cortical tissue in psychiatric diseases was obtained from previously published data (Gandal et al., 2018). To infer which neuron subtypes contributed to the previously reported decrease in neuronal gene modules in ASD, SCZ, and BD we decided to only consider genes that were likely to be expressed by neurons. To do this we first defined a set of 1414 “neuron genes” as those that had higher expression in neurons compared with all other brain cell types (astrocytes, microglia, endothelial cells, oligodendrocytes) in each of two distinct datasets (GSE52564 and GSE71585). We then took these neuronal genes and looked at their expression in our dataset of neuron subtypes to determine which of the four neuron subtypes (EXC, PV, SST, or VIP) had the highest expression of each gene. From this we calculated the background proportion of subtype-specific genes for this initial set of neuron genes. Next, we overlapped the list of genes downregulated in a particular psychiatric disease with our 1414 neuron genes, and performed the same analysis across the neuron subtypes data to determine the proportion of subtype-specific genes for each disease. To determine the significance of the results in each disease, we calculated empirical p values. Briefly, for a given set of N neuron genes downregulated in a disease, we performed 1000 rounds of sampling N genes (without replacement) from the 1414 set of background neuron genes, and then calculated the proportions of subtype-specific genes for each sampling. Empirical p values were then calculated based on the frequency of observing a proportion as high (or low) as observed in the disease downregulated neuron genes, over the 1000 rounds of sampling. As a control the same analysis was applied to genes found downregulated in inflammatory bowel disease (IBD) patients, a gene set that is unlikely to contain meaningful changes in brain neuron genes, and was previously found to not correlate with the ASD, SCZ, and BD brain differential expression datasets (Gandal et al., 2018).
Comparison of neuronal gene expression to psychiatric GWAS datasets.
Genome-wide association study (GWAS) hits for ASD, BD, and SCZ were downloaded from the NHGRI-EBI GWAS catalog (https://www.ebi.ac.uk/gwas/home) November 1, 2018 and included multiple studies for each disease. GWAS hits were filtered for adjusted p ≤ 5e−08. For each significant SNP in the GWAS data, we took the assigned genes as reported by the data tables from the GWAS catalog. The reported genes for the GWAS hits were then overlapped with the 1414 neuron genes and downstream analysis was performed as for the transcriptomics datasets.
Accessions.
Gene expression omnibus: GSE122100.
Results
Differential gene expression in major neuron subtypes
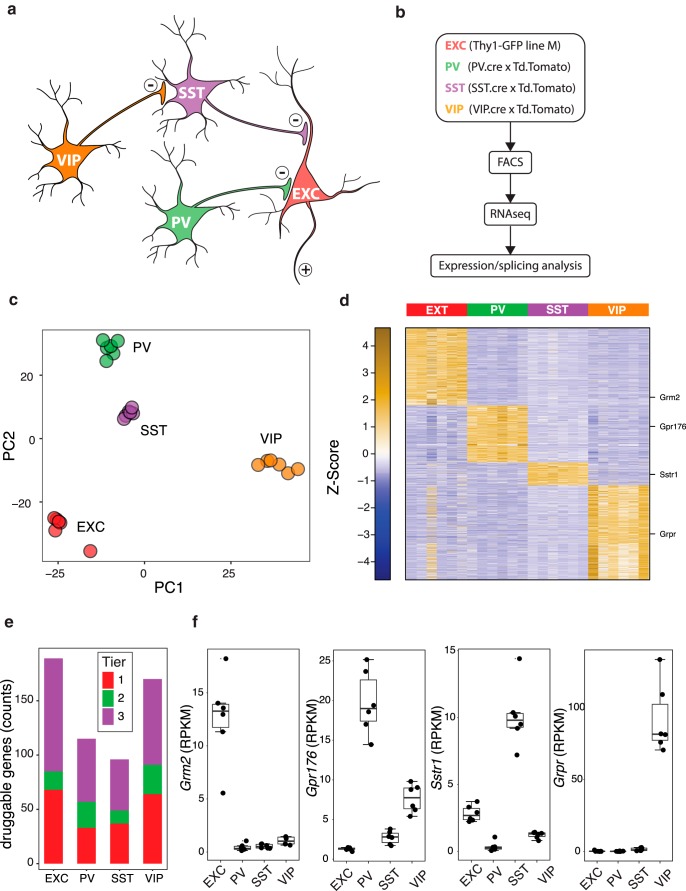
We used FACS to purify a subset of EXC neurons (Thy1-GFP M line), and PV, SST, and VIP interneurons (using Cre driver lines crossed to a tdTomato reporter line; see Materials and Methods) from the entire cortex and hippocampus of the respective mouse lines (Fig. 1a,b, Fig. 1-1). By using the Thy1-GFP M line the EXC populations are a mixture of cortical pyramidal neurons (layer 5), dentate gyrus granule cells, and CA1–3 pyramidal neurons. Principal component analysis of RNA-seq gene expression for the samples from each animal (n = 6 mice/neuron subtype) showed strong clustering and clear separation by cell type (Fig. 1c). We identified genes as enriched in a particular neuron subtype if expression was significantly higher [log2(fold-change) ≥ 1, adjusted p ≤ 0.05] in that subtype by pairwise comparisons to each of the other three subtypes. Overall 2403 genes were enriched in a neuron subtype-specific manner (738 EXC, 544 PV, 219 SST, 902 VIP; Fig. 1d, Fig. 1-2).
Figure 1.
RNA-seq analysis of cortical neuron subpopulations. a, Simplified diagram of a cortical circuit including EXC pyramidal neurons, and three classes of inhibitory interneurons: PV, SST, and VIP neurons. b, Experimental scheme: four lines of transgenic mice were used with fluorescent proteins expressed in one of the four neuron subtypes; cortex and hippocampus were dissociated, and the fluorescent cells FACS purified for transcriptome profiling by RNA-seq. c, Principal components analysis of the 24 samples, based on overall gene expression, demonstrating clear separation of clusters by neuron subtype. d, Heatmap of z-score normalized RPKM expression levels of genes significantly enriched in each neuron subtype. e, Counts of genes specifically enriched in each neuron subtype with “druggable genes” in Tier 1 (targets of approved small molecules and biotherapeutic drugs and clinical-phase drug candidates), Tier 2 (targets with known bioactive drug-like small-molecule binding partners as well as those with ≥50% identity with approved drug targets), and Tier 3 (genes encoding secreted or extracellular proteins, proteins with more distant similarity to approved drug targets, and members of key druggable gene families not already included in Tiers 1 or 2, such as GPCRs, nuclear hormone receptors, ion channels, kinases, and phosphodiesterases). f, Specific examples of druggable genes enriched in each neuron subtype, visualized as boxplots of RPKM expression levels. Boxes show median and interquartile ranges, and whiskers show full data without outliers. Genes are also indicated on the heatmap in d.
RNA-seq quality metrics RNA-sequencing quality control metrics across the 24 samples generated in this study (a). Samples are in columns. QC metrics are shown in rows. Rows contain different quality control metrics used to assess each sample. Descriptions of quality control metrics are indicated by text labeling each row. 5’/3’ coverage plots for the current study (b) and the samples from GSE77243 (c) and GSE71585 (d) included for comparison in our website resource. Briefly, for each gene g a coverage vector cgi was calculated giving the number of reads overlapping position i of the gene, for i=1...ng tracking the annotated exonic bases of the gene from 1 to the total number of exonic bases ng of gene g, from 5’ (i = 1) to 3’ (i=ng) in the direction of transcription. Then the gene was divided into 20 equally spaced bins Bb of ng/20 bases each. The average coverage acgb was then calculated as the average over all the bases in the bin of cgi, or acgb=1/(ng/20) ∑iEBb cgi . Next, these average coverages acgb were converted into normalized coverages ncgb by dividing by the average coverage over the entire gene: acg=1/ng ∑(i=1)(ng)cgi and then ncgb=acgb/acg . This normalized coverage ncgb is then a matrix whose rows are all the genes and columns are the twenty bins. For speed of calculation and robustness, we selected 500 genes at random among those that were expressed with at least 4 nRPKMs in at least 1/4 of the samples. Finally, we calculated the median normalized coverage for each bin over those 500 genes: ncb=“median”gE“500 genes” (ncgb). This statistic is plotted as the Median Relative Coverage. Download Figure 1-1, PDF file (624KB, pdf)
Differential gene expression Differential expression results per gene, including average RPKM expression levels per neuron subtype per gene, and neuron subtype pairwise log2FC and p-value statistics. Download Figure 1-2, PDF file (624KB, pdf)
Druggable targets in single neuron subtypes Druggable targets enriched in each neuron subtype, including druggable genome annotations. Download Figure 1-3, PDF file (624KB, pdf)
Druggable targets in pairs of neuron subtypes Druggable targets enriched in pairs of neuron subtypes, including druggable genome annotations. Download Figure 1-4, PDF file (624KB, pdf)
Drug targets enriched in neuron subtypes
Genes selectively expressed in particular neuron subtypes could represent potential targets for manipulating specific aspects of circuit function. We therefore compared the list of differentially expressed genes to the druggable genome (Finan et al., 2017; Fig. 1e). This list includes Tier 1 (targets of approved small molecules and biotherapeutic drugs and clinical-phase drug candidates), Tier 2 (targets with known bioactive drug-like small-molecule binding partners as well as those with ≥50% identity with approved drug targets), and Tier 3 (genes encoding secreted or extracellular proteins, proteins with more distant similarity to approved drug targets, and members of key druggable gene families not already included in Tier 1 or 2, such as GPCRs, nuclear hormone receptors, ion channels, kinases, and phosphodiesterases). This analysis identified 189 druggable targets enriched in EXC neurons and 115, 96, and 170 targets enriched in PV, SST, and VIP interneurons, respectively (Fig. 1-3). In addition, 1446 more druggable genes were selectively enriched in two of the four cell types (Fig. 1-4). These targets include voltage- and ligand-gated ion channels, enzymes, GPCRs and others. This indicates the potential for selectively modulating specific neuron subtypes or combinations of neuron subtypes via targeting of these druggable proteins. Examples of GPCRs with selective expression include Grm2 (metabotropic glutamate receptor 2) in EXC neurons, Gpr176 (orphan GPCR) in PV neurons, Sstr1 (somatostatin receptor type 1) in SST neurons, and Grpr (gastrin-releasing peptide receptor) in VIP neurons (Fig. 1f). Mining the list of >2000 identified classically druggable genes with neuron subtype-specific expression patterns should facilitate the identification of drug targets for manipulating circuit function.
Neuron subtype-specific alternative splicing
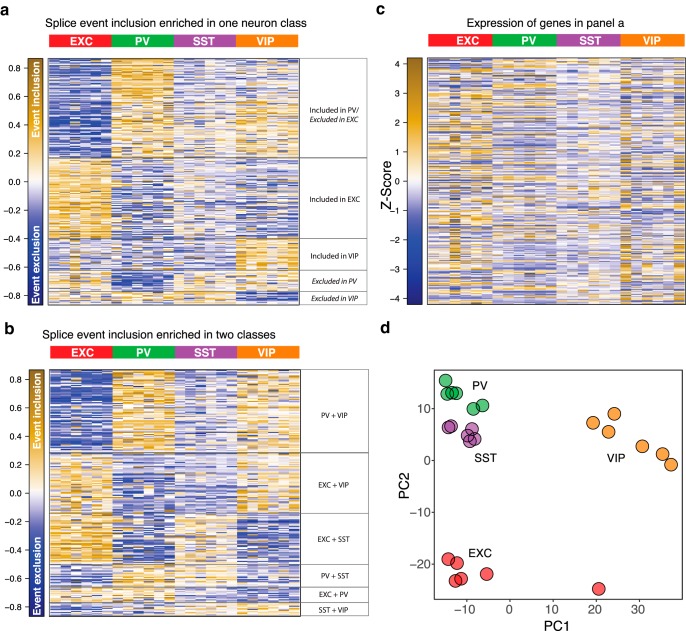
Although alternative splicing can have important functional impact in the brain (Raj and Blencowe, 2015), genome-wide analysis of differential splicing using junction reads has not been extensively performed. Whereas previous work has examined the alternative splicing programs of developing SST and PV neurons (Wamsley et al., 2018), here we use splice graph analysis to detect and quantify junction reads and assess differential usage of splice variants across adult EXC, PV, SST and VIP neurons. To comprehensively evaluate splicing variation, we tested for any splicing event that occurred significantly more frequently [|log2(fold-change)| > 0.32, adjusted p ≤ 0.05] in one neuron subtype compared with all of the other three. This analysis found many splice events that were enriched in a single neuron subtype (Fig. 2a, Fig. 2-1). The largest cluster consisted of splicing events both significantly enriched in PV neurons and significantly depleted in EXC neurons (n = 124), indicating that EXC and PV neurons have the most dissimilar splicing activity of the neuron classes. The next largest cluster consisted of EXC-enriched events (n = 100), followed by smaller clusters that showed VIP enrichment (n = 39), PV depletion (n = 26), and VIP depletion (n = 11). We also tested for splice events selectively occurring in only 2 of the 4 neuron classes and found 231 additional selective splice events (Fig. 2b, Fig. 2-2). Notably, the genes showing the most differential splicing did not correlate strongly with the genes that were differentially expressed across the neuron subtypes (Fig. 2c). At the same time, PCA based on splice usage clearly separated the cell types (Fig. 2d). Thus, differential splicing is used as an additional layer of specification of neuron subtype diversity that is independent from differential gene expression.
Figure 2.
Neuron subtype-specific gene splicing. a, Heatmap of splice event variant inclusion frequencies (centered to the mean per variant across samples) across the samples, for events which were significantly differentially spliced in one neuron subtype compared with the other three. General descriptions of the patterns are included on the right. b, Heatmap, as in a, for splice events significantly differentially spliced in two neuron subtypes. c, Heatmap of z-score normalized RPKM expression levels of the same genes which contain the splice events in a, preserving the ordering of rows as in a. d, Principal components analysis of the 24 samples, based on the overall splice events quantification, demonstrating clear separation of clusters by neuron subtype.
Differentially spliced events in single neuron subtypes Differentially spliced events in a single neuron subtype and corresponding log(fold change) and p-value statistics. Variant types are defined as follows: SE (skipped exon), S2E (two consecutive exons skipped), RI (retained intron), MXE (mutually exclusive exons), A5SS (alternative 5’ splice site), A3SS (alternative 3’ splice site), AFE (alternative first exon), ALE (alternative last exon), AS (alternative start other than AFE), AE (alternative end other than ALE). Download Figure 2-1, PDF file (1.9MB, pdf)
Differentially spliced events in pairs of neuron subtypes Differentially spliced events in two neuron subtypes and corresponding log(fold change) and p-value statistics. Variant types are defined as in Figure 2-1. Download Figure 2-2, PDF file (1.9MB, pdf)
Differential splicing of voltage and ligand-gated ion channels
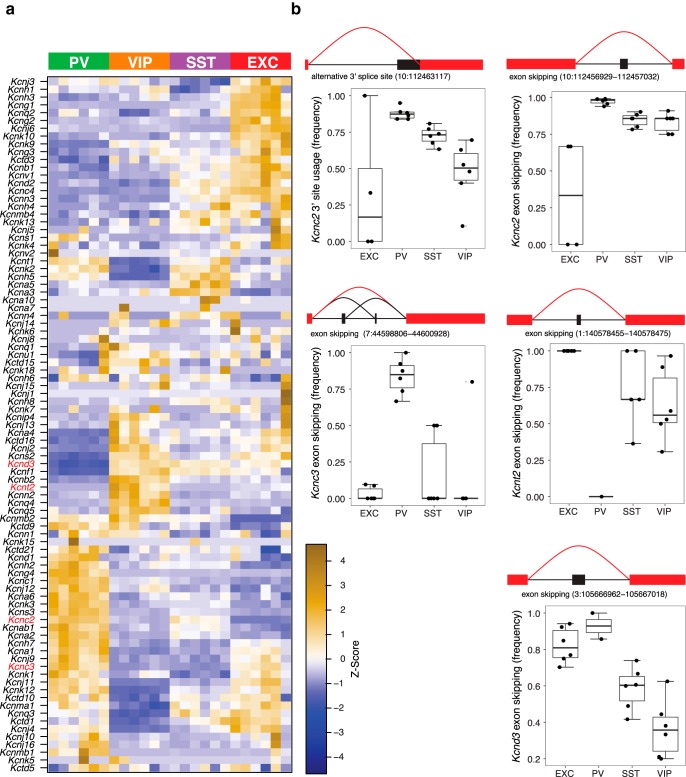
Potassium channels provide an example of the combination of differential expression and differential splicing. In addition to neuron subtype-specific expression patterns (Fig. 3a), we observed many examples of differential splicing (Fig. 3b). For example, Kcnc2, which encodes Kv3.1 channels, is alternatively spliced at two distinct sites, with PV and EXC neurons having the most different exon usage, and SST and VIP neurons exhibiting somewhat intermediate profiles at both sites. Kcnc3, which encodes Kv3.3 channels, exhibits PV-selective skipping of two adjacent exons, and Kcnt2, which encodes KNa channels, also exhibits PV-selective exon skipping. For Kcnd3, which encodes Kv4.3 channels, SST and VIP neurons show contrasting exon inclusion compared with EXC and PV neurons. Although less is known about functional impacts of Kv4.3 and KNa alternative splicing in neurons, Kv3.1b is implicated in axonal localization, and Kv3.1b and Kv3.3b isoforms are associated with the capacity for repetitive spiking (Gu et al., 2012; Veys et al., 2013), suggesting differential potassium channel splicing could contribute to neuron subtype functional specialization.
Figure 3.
Differential exon usage and expression of potassium channels. a, Differential expression of potassium channel genes, depicted as a heatmap of z-score normalized RPKM gene expression values. b, Examples of potassium channel differential splicing are illustrated for Kcnc2 (two distinct sites), Kcnc3, Kcnt2, and Kcnd3. Genomic locations of the splicing events are indicated. Boxplots depict the frequency of the indicated splicing events. Boxes show median and interquartile ranges, and whiskers show full data without outliers.
We also observed cell-type-specific inclusion of exons with known functional importance in ligand-gated ion channels including NMDA, AMPA, and GABAA receptors (Fig. 4a–c, Fig. 4-1). Alternative splicing of Grin1 modulates NMDAR functional properties, with inclusion of exon 5 resulting in reduced proton inhibition and a faster deactivation rate (Traynelis et al., 1995; Rumbaugh et al., 2000; Regan et al., 2018). We found that exon 5 is highly excluded from Grin1 in excitatory neurons, but more often included in interneurons, especially PV neurons (Fig. 4a). For AMPARs we observed neuron subtype-specific usage of the alternative exons 15 (flip) and 14 (flop) of Gria2, which modulate pharmacology and deactivation kinetics (Sommer et al., 1990). Whereas EXC and SST neurons used a mixture of flip and flop isoforms, PV neurons, and to a lesser extent VIP neurons, almost exclusively express the flop isoform (Fig. 4b). For GABAA receptors we observed differential splicing of the γ2 subunit gene, Gabrg2, which can include or exclude a region in the cytoplasmic loop between transmembrane domains 3 and 4 (resulting in the γ2L or γ2S isoforms, respectively; Whiting et al., 1990). PV neurons almost always expressed GABRG2-γ2L, whereas other neuron subtypes, especially excitatory neurons more often used GABRG2-γ2S (Fig. 4c). As γ2L has been associated with the capacity for synaptic localization of GABAARs (Meier and Grantyn, 2004), this suggests alternative splicing could modulate the synaptic function of certain GABAARs in a neuron subtype-specific manner.
Example sequencing coverage at splice junctions IGV genome browser screenshots of the sequencing coverage at and surrounding the splice junctions featured in Figure 4. In the genome browser screenshots, each row represents the read coverage of a single sample. The first 6 red samples are the EXC samples, followed by 6 green PV samples, then 6 purple SST samples, and 6 orange VIP samples. Read counts of splice junctions corresponding to the four events featured in Figure 3 (e-h). In the splice read count plots, each row shows counts of reads by sample for a specific neuronal population isolated. Read counts reflect the total count of reads supporting and spanning a splice event variant, as indicated in methods. Gene models were then mapped to the same genomic coordinates for display in the figure. Each sample is represented with a different colored line as indicated in the key to the right of each row. In the bottom panel, exons are indicated by boxes and introns by lines. Download Figure 4-1, XLS file (15.2MB, xls)
An interesting illustration of differential splicing is the Cacna1a gene (Cav2.1 calcium channel), which is implicated in migraine, epilepsy and episodic ataxia, and undergoes functionally important alternative splicing at exon 37 (Bourinet et al., 1999). We found that while EXC neurons mostly used exon 37b, interneurons more often used exon 37a, with PV interneurons exclusively using exon 37a (Fig. 4d). A recent study showed that exon 37 can control short-term synaptic plasticity in primary neurons, with exon 37b specifying short-term facilitation, and exon 37a specifying short-term depression (Thalhammer et al., 2017). Our differential splicing data would predict that in contrast to facilitation in pyramidal neurons, PV interneurons should exhibit short-term depression. To investigate this, we used optogenetic activation of PV interneurons in brain slices and recorded postsynaptic responses in EXC neurons (Fig. 4e). PV to EXC synapses showed pronounced short-term depression with high-frequency stimulation, as predicted by exon 37a usage by PV neurons (Fig. 4f). In contrast, stimulation of the EXC inputs produced short-term facilitation of postsynaptic responses in EXC neurons, consistent with exon 37b usage. This correlation of exon 37 usage with short-term plasticity phenotypes should motivate future experiments using neuron subtype-specific manipulation of exon 37 to test for a potential causal role of exon 37 splice variant usage in cell-type-specific short-term plasticity.
Comprehensive analysis of neurexin exon usage
The interaction of presynaptic neurexins with postsynaptic neuroligins is critical to synapse development, and alterations in the genes encoding these proteins are linked to neurodevelopmental disease including ASD (Südhof, 2008). Neurexins (Nrxn1–3), are expressed in long (α) or short (β) forms, and have six canonical alternative splice sites (AS1–6), leading to thousands of potential spliceforms that can be expressed in the brain (Schreiner et al., 2014; Treutlein et al., 2014; Fig. 5a). Neurexin alternative splicing is critical in determining synaptic connectivity (Chih et al., 2006; Futai et al., 2013; Nguyen et al., 2016). For example, AS4 exclusion versus inclusion can be important for specifying GABAergic versus glutamatergic synapse in the context of β-neurexins (Chih et al., 2006). Previous studies comparing pairs of neuron subtypes or probing electrophysiologically defined neuron subtypes have found examples of neuron subtype-specific neurexin isoform usage and have provided evidence for a specific synaptic adhesion code (Fuccillo et al., 2015; Földy et al., 2016; Nguyen et al., 2016). Our work now provides comprehensive information on the splice form usage at all splice sites (AS1–6) in Nrxn1–3, across EXC, PV, SST, and VIP neurons (Fig. 5).
Figure 5.
Comprehensive analysis of neurexin canonical splicing. a, Gene models of Nrxns1–3, with canonical alternative splicing regions (AS1–6) in red, and the start of the β isoforms in black. b, Quantification of α isoform exon expression evidence (as a proxy for α–β isoform ratios). c, Frequency of exon inclusion for AS1–6 exons across each neuron subtype. d, Validation of AS3 differential splicing using semiquantitative RT-PCR analyses of Nrxn1, Nrxn2, and Nrxn3, using primers that surround the respective AS3 exons. Images show example gels depicting the shift in PCR product length for VIP samples relative to EXC, SST, and VIP samples. Predicted product lengths are as follows: nrxn1 204 bp with exon, 177 bp without; Nrxn2 204 bp with exon, 165 bp without; nrxn3 177 bp with exon, 138 bp without. Plots show relative proportion of detected inclusion product. Bar charts show mean ± SEM (n = 6 EXC, 6 PV, 2 SST, and 3 VIP). e, Expression of genes encoding neurexin interacting proteins as a z-score normalized heatmap of RPKM expression levels (left), and boxplots of four examples enriched or depleted in 1–2 neuron subtypes (right). *Exon inclusion in an indicated cell type is significantly different from each of the other three neuron subtypes (two-sided t test, p < 0.05).
Differential expression of α versus β neurexins in the neuron subtypes is evident in the ratio of α unique exon reads to α/β common reads (Fig. 5b). For example, VIP neurons express the highest relative amount of the α transcript of Nrxn2, whereas EXC neurons significantly more often express β transcripts. In addition to greater inclusion of AS4 in PV neurons versus EXC neurons, our data reveals distinct usage of this exon in the other neuron types across the different neurexins (Fig. 5c). Interestingly, for Nrxn1 and Nrxn3, SST and VIP neurons include AS4 at similarly high levels as in PV neurons, whereas for Nrxn2, SST and VIP neurons include AS4 at the lower level of EXC neurons. Our data also reveals significant differential usage of other alternative splice sites with a striking example being the VIP neuron-specific exclusion of AS3 of Nrxn1, Nrxn2, and Nrxn3, which we confirmed by PCR (Fig. 5c,d). Combined with the neuron subtype expression patterns of various neurexin interacting proteins (Fig. 5e), this neurexin splicing information provides a roadmap for dissecting the complex functional roles of different isoforms in specifying neuronal connectivity patterns.
A searchable web resource for comparing differential expression and splicing across datasets
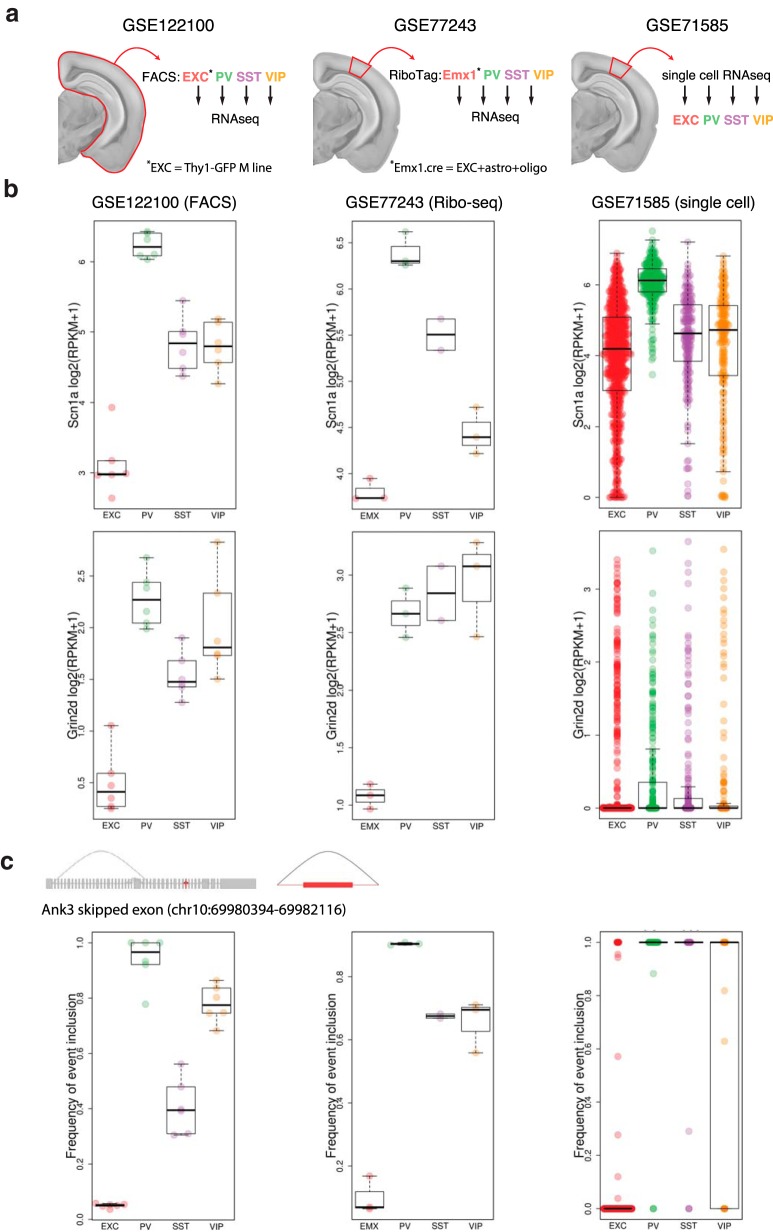
The examples highlighted above represent only a small sampling of our neuron subtype-specific transcriptomic data. To provide an accessible and comprehensive resource, we have created a searchable web interface including expression data for all genes and analysis of every splice event that varied significantly across neuron subtypes (http://research-pub.gene.com/NeuronSubtypeTranscriptomes). This resource includes information on both protein-coding genes and non-coding RNAs. In addition, we have imported two other datasets that contain RNA-seq data from EXC, PV, SST, and VIP neurons (Mardinly et al., 2016; Tasic et al., 2016), and applied our analysis pipeline to these additional samples. Analysis of these other datasets is provided alongside our data to allow assessment of the consistency of differential expression and splicing across experiments. These datasets relating to neuron cell-type-specific RNA expression were collected by different approaches; (1) FACS-sorted neurons from cortex + hippocampus of fluorescent reporter mice, each sample being an individual mouse of either sex, with Thy1-GFP M line representing EXC neurons (present study; GSE122100); (2) ribosome-associated RNA purified from visual cortex, each sample being a pool of three mice including both sexes, using Emx-Cre for EXC neurons (GSE77243; Mardinly et al., 2016; which due to developmental Emx expression will also include astrocytes and oligodendrocytes; only samples from mice raised under control conditions were included for our analysis); and (3) single-cell RNA-seq obtained from dissociated visual cortex in male mice (GSE71585; Tasic et al., 2016), with EXC, PV, SST, and VIP data pooled from subclasses of these cell types for our analysis (Fig. 6a).
Figure 6.
Example expression and splicing data from the web interface. a, In addition to the present dataset (GSE122100) we provide analysis of two other datasets. Whereas GSE122100 used entire cortex (including hippocampus), GSE77243 and GSE71585 used only visual cortex. GSE77243 used Ribo-tag purification of neuron subtypes for RNA-seq, including Emx1.cre to isolate EXC neurons (which, due to developmental expression, will also include astrocytes and oligodendrocytes). GSE71585 used single-cell RNA-seq, and we pooled these data at the level of broad neuron subtype classes for analysis. b, Example expression data are shown for genes with relatively high expression (Scn1a) and relatively low expression (Grin2d). c, Example splicing quantification is shown using the gene Ank3 focusing on the skipped exon event at chr10:69980394-69982116.
By providing simultaneous analysis of multiple datasets this new resource allows assessment of the consistency of observations of differential expression and splicing. Thus, replication of a differential transcriptomic event in one dataset can be evaluated by confirmation in the other datasets, providing more confidence than just relying on statistical significance in a single dataset. Figure 6b shows example expression plots from all three datasets as they appear on the web interface, using the genes Scn1a and Grin2d, which represent examples of high and low neuronal expression levels, respectively (Fig. 6b). For Scn1a, all three datasets agree fairly well with lowest expression in EXC neurons, highest expression in PV neurons, and intermediate expression levels in SST and VIP neurons. However, in the case of Grin2d, effects of dropout in the single cell dataset make it hard to resolve the higher expression in interneuron subtypes compared with EXC neurons that is evident in the other datasets. Figure 6c shows example splice event inclusion plots as they appear on the web interface using the gene Ank3, which is associated with BD and SCZ, with isoform-specific alterations associated with modified disease risk (Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011; Sklar et al., 2011; Rueckert et al., 2013; Hughes et al., 2016, 2018). A significant differential splicing event observed for Ank3 in our dataset involving greater usage of an internal exon by PV and VIP neurons compared with EXC and SST neurons is depicted in Figure 6c. Although both of the other datasets agree with our observations of very low exon inclusion in EXC neurons and very high inclusion in PV and VIP neurons, the frequency of exon inclusion for SST and VIP neurons varies across the datasets. The discordant observation of 100% event inclusion in PV and SST neurons in the single cell dataset is likely due to under-sampling, because >97% of the samples with exon reads had either 0 or 1 read. Overall, despite differences in methods of isolating and sequencing cell-type-specific mRNAs all three datasets showed significant correlations in the observed profiles of gene expression and exon usage (Fig. 7a,b).
Figure 7.
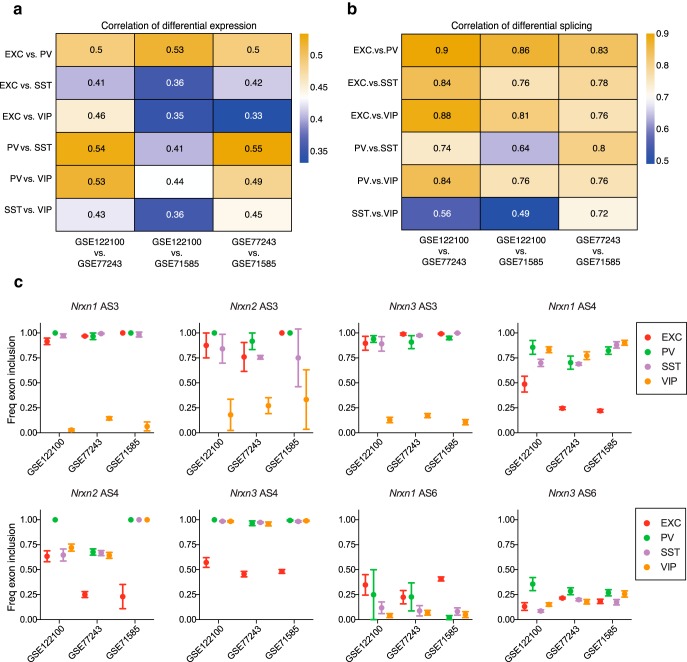
Correlation of gene expression levels and splicing across datasets. a, b, Correlations between each pairwise neuron subtype comparison, across the three RNA-seq datasets. Correlations of log2(fold-change) expression levels for all genes (a) and splice variants (b). For each pair of cell types (rows), we extracted the vectors of log2 fold-changes for all genes. Then, for each pair of datasets (columns) we calculated the Pearson correlations between these vectors. b, A similar analysis for splice variants instead of genes, but only including splice variants which were significantly differentially spliced in at least one of the three datasets [|log2(fold-change)| > 0.32, adjusted p ≤ 0.05]. c, Plots show the relative exon inclusion for Nrxn1, Nrxn2, and Nrxn3 for AS3, AS4 and AS6 in the three datasets. Plots show mean ± SEM.
Although our splice junction analysis pipeline was previously validated for accurately predicting differential splicing in neurons (Srinivasan et al., 2016), comparison across datasets allows further validation of differentially expressed events. For example, analyses of the three datasets largely agree in terms of subtype-specific neurexin splice events (Fig. 7c). In particular the VIP exclusion of AS3 in Nrxn1–3 seen in our dataset is also robustly observed in the other datasets, and the profiles of AS4 and AS6 splicing are also largely replicated. In the case of Nrxn2 AS4, the other datasets agree with relatively higher PV versus EXC inclusion, but they differ from our dataset in that SST and VIP neurons also exhibit relatively higher inclusion of AS4. This could potentially be due to brain region-dependent splice usage by SST and VIP neurons, as our dataset includes hippocampal neurons, while the other datasets are focused on visual cortex. In addition to generally good agreement across our analysis of RNA-seq datasets, the validity of our splice predictions is supported by data from other approaches. For example, in addition to our PCR confirmation of AS3 differential splicing (Fig. 5d), PCR analysis following ribo-trap purification of hippocampal CamK2(+) and PV(+) cells(Nguyen et al., 2016) agrees with the differential splicing across neurexins that we see for AS4 and AS6 between EXC and PV neurons. In addition, a recent single-cell analysis comparing Nrxn1 and Nrxn3 splicing in pyramidal cells versus Prox1(+) versus Lhx6(+) interneurons (Lukacsovich et al., 2019) agrees with our identification of differential splice events.
Using neuron subtype transcription patterns to analyze human datasets
The transcriptomic profiles of interneuron classes can be useful in interpreting expression data from human patient brain tissue. For example, a recent network analysis of psychiatric disorder patient cerebral cortex samples found that while expression of an astrocyte gene module was increased in ASD, SCZ, and BD, neuronal gene modules had decreased expression in these disorders (Gandal et al., 2018). Although the previous analysis did not consider neuron subtypes, we can apply our data on neuron subtype-enriched gene expression to dissect the psychiatric patient dataset with greater resolution (Fig. 8a). Compared with the expected background levels of neuronal genes, or to a control population (IBD), we observe that a greater than expected proportion of the neuronal genes downregulated in cortical tissue of ASD and SCZ patients is enriched in PV neurons, while in BD a greater than expected proportion is enriched in EXC neurons (Fig. 8b). In principle this could reflect either a loss of PV neurons in ASD and SCZ and a loss of EXC neurons in BD, and/or a decrease in signature gene expression in these cell types in these diseases. Conversely, the lower than expected proportion of downregulated genes in ASD, SCZ, and BD that are enriched in VIP neurons could reflect either a relative overabundance of VIP neurons or a misregulation of signature gene expression in VIP neurons in these psychiatric diseases. We also applied this type of analysis to gene sets implicated in ASD, SCZ, or BD by GWASs (Fig. 8c). Consistent with the neuron subtypes implicated by analysis of cortex gene expression, this analysis showed trends toward enrichment of BD GWAS genes in EXC neurons and ASD and SCZ GWAS genes in PV neurons. At the same time, distinct from the ASD brain expression analysis, ASD GWAS genes were significantly enriched in EXC neurons and depleted in SST neurons.
Figure 8.
Use of neuron subtype expression patterns to analyze psychiatric patient brain gene expression changes. a, Data analysis work flow. Genes that were most highly expressed in neurons compared with all other brain cell types in two datasets (GSE52564 and GSE71585) were included in the background neuron-gene set. Neuron genes were then identified in the set of genes downregulated in psychiatric patient brains (Gandal et al., 2018). The proportion of genes enriched in each neuron subtype from our dataset was then determined for the background gene set and each psychiatric disease diagnosis. b, Distribution of neuronal genes most highly expressed in each neuron subtype, and overlapped with genes downregulated in brain tissue from ASD, SCZ, and BD patients or an IBD downregulated gene set as a control. The frequency of disease downregulated genes across neuron subtypes compared with all neuronal genes was determined and empirical p values from n = 1000 samplings were calculated [***empirical p < 0.001, indicating the observed value is significantly more (or less) frequent than the background level for all neurons]. c, Analysis of psychiatric patient GWAS genes sets using neuron subtype expression patterns. GWAS hits for ASD, BD, and SCZ were downloaded from the NHGRI-EBI GWAS catalog (https://www.ebi.ac.uk/gwas/home) November 1, 2018 and included multiple studies for each disease. GWAS hits were filtered for adjusted p ≤ 5e−08. The reported genes for the GWAS hits were then overlapped with the 1414 neuron genes and downstream analysis was performed as in Figure 8, a and b, with the GWAS genes, instead of genes downregulated in psychiatric disease. Asterisk symbol denotes an empirical p < 0.05.
Discussion
We have identified >7000 genes (comprising ∼80% of genes expressed with mean RPKM ≥ 2 in any neuron subtype) that have differential expression and/or splicing between EXC, PV, SST, and VIP neuron subtypes. This transcriptomic information provides insight into the basis for functional specialization of the principal classes of interneurons and indicates many druggable targets with neuron subtype enrichment, which could theoretically allow manipulation of circuit function. For example, targets that could be manipulated to enhance activation of PV and/or SST neurons could help suppress overall circuit activity. This could be beneficial in epilepsy and other diseases with dysfunctional inhibition and network hyperactivity such as SCZ and AD. Conversely, targets that could be used to suppress activation of EXC and/or VIP neurons could have similar benefits through decreasing excitation directly or by reducing dis-inhibition. In identifying druggable targets we included genes that were significantly enriched in one neuron subtype, but not necessarily exclusively expressed in that subtype [log2(fold-change) ≥ 1, adjusted p ≤ 0.05]. As these criteria will exclude some genes with modest but functionally important neuron subtype enrichment, and considering the expanding repertoire of targets that are druggable, our list of neuron class-specific targets represents a conservative estimate of gene products that could potentially be manipulated to influence circuit function. Other caveats to be kept in mind are that purification of cells by FACS can alter expression of some activity-dependent genes (Wu et al., 2017) and that protein expression of potential drug targets may differ from the RNA expression levels measured here.
Our analysis of transcript splicing indicates subclasses of neurons not only use differential gene expression, but also independently use differential splicing to achieve functional specialization. Our comprehensive analysis expands on previous reports of differential splicing of specific genes in specific cell types, and provides extensive novel information on differential splicing across neuron classes. For example, it was previously reported that in contrast to EXC neurons, PV neurons included AS4 of neurexins (Nguyen et al., 2016). Our comprehensive analysis reveals unique usage patterns of AS4 of Nrxn1, Nrxn2, and Nrxn3 across EXC, PV, SST, and VIP neurons, as well as unique usage of other splice sites including VIP-selective exclusion of AS3 in all three neurexins (Fig. 5). In another example we find novel differential splicing in Ank3 across interneuron types, especially PV and VIP, compared with EXC neurons (Fig. 6c), which supports potential roles for these interneuron subtypes in BD and SCZ pathophysiology. Overall the comprehensive picture of gene expression and exon usage provided here should provide a foundation for exploring the complex physiological and pathological roles of various genes in circuit function. It should also be noted that there are probably many differential splicing events that are not detected by our present analysis because of limitations including the nature of the RNA collection protocols, read depth in the datasets, and relatively sparse coverage of 5′ ends of genes.
The website (http://research-pub.gene.com/NeuronSubtypeTranscriptomes) can be used to compare differential expression and splicing across our FACS dataset, the ribo-seq dataset, and the single-cell dataset. As each dataset has distinct advantages and limitations, this should allow for evaluation of the robustness of any neuron subtype-specific expression or splicing result. We anticipate that this resource will be used as a starting point for exploring the potential physiological roles and the therapeutic potential of genes that could affect neuronal and circuit function. These data will be particularly useful when assessed in the context of other available datasets, which provide complementary information. To provide high power for analysis of expression and differential splicing, we have focused on several broad neuron classes (EXC, PV, SST, VIP). Upon identifying a gene with an interesting transcriptomic profile in one of these broad neuron classes, it is informative to examine that gene across the subclasses of EXC, PV, SST, and VIP neurons as well as other types of inhibitory neurons that have been identified in single cell datasets (Zeisel et al., 2015; Tasic et al., 2016; Paul et al., 2017; Hrvatin et al., 2018). Similarly, in considering potential drug targets, it will be important to look within these single cell datasets and other datasets that allow comparison of neuronal and non-neuronal expression, as well as expression across brain regions, and in comparisons to other tissues (Zhang et al., 2014; Sharma et al., 2015). For example, although we pooled entire cortex and hippocampus with the goal of identifying broadly consistent transcriptomic events that could be amenable to targeting with drugs, other datasets better speak to region-specific differential events.
Combining data on brain cell taxonomy and disease-associated gene sets has the potential to reveal key cell types involved in pathophysiology. For example, this approach has identified neuron types in different brain regions as potentially underlying pathophysiology in diseases including SCZ and autism, including cortical interneurons in general (Ben-David and Shifman, 2012; Xu et al., 2014; Chang et al., 2015; Skene et al., 2018). In this context, our robust dataset on cortical interneuron subtype transcriptomic signatures will be especially valuable in dissecting the roles for interneuron subtypes. We illustrate this type of usage of our data by mapping neuron subtype gene expression profiles onto the sets of neuronal genes recently found to be decreased in ASD, SCZ, and BD brains (Gandal et al., 2018). For example, this analysis specifically implicates PV neuron loss/dysfunction in ASD and SCZ while implicating EXC neuron loss/dysfunction in BD (Fig. 8b). Analysis of neuronal genes from ASD, SCZ, and BD GWAS studies using our dataset further supports a role for these neuron subtypes in these disorders (Fig. 8c). Interestingly, the analysis of ASD GWAS genes found significant enrichment in EXC neurons and depletion in SST interneurons, even though these cell types were not implicated in the analysis of downregulated genes from ASD patient brains. This emphasizes that the neuronal populations involved in conferring risk for a disease are not necessarily the same populations that exhibit alterations during manifestation of that disease. In general, future efforts studying dysfunction of specific interneuron subclasses within subsets of patients could not only help reveal the roles of circuit dysfunction in neurological disease, but could also be useful in stratifying patients within a disease diagnosis as likely to benefit from therapeutics targeting specific aspects of circuit function.
Footnotes
We thank Zora Modrusan for help with library preparation and RNA sequencing.
Conflict-of-interest: all authors are current or former employees of Genentech.
References
- Anders S, Reyes A, Huber W (2012) Detecting differential usage of exons from RNA-seq data. Genome Res 22:2008–2017. 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos M, Vida I, Jonas P (2007) Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci 8:45–56. 10.1038/nrn2044 [DOI] [PubMed] [Google Scholar]
- Ben-David E, Shifman S (2012) Networks of neuronal genes affected by common and rare variants in autism spectrum disorders. PLoS Genet 8:e1002556. 10.1371/journal.pgen.1002556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourinet E, Soong TW, Sutton K, Slaymaker S, Mathews E, Monteil A, Zamponi GW, Nargeot J, Snutch TP (1999) Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nat Neurosci 2:407–415. 10.1038/8070 [DOI] [PubMed] [Google Scholar]
- Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI (2009) Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 459:663–667. 10.1038/nature08002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D (2015) Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 18:191–198. 10.1038/nn.3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P (2006) Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron 51:171–178. 10.1016/j.neuron.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR (2000) Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28:41–51. 10.1016/S0896-6273(00)00084-2 [DOI] [PubMed] [Google Scholar]
- Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J, Galver L, Kelley R, Karlsson A, Santos R, Overington JP, Hingorani AD, Casas JP (2017) The druggable genome and support for target identification and validation in drug development. Sci Transl Med 9:eaag1166. 10.1126/scitranslmed.aag1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földy C, Darmanis S, Aoto J, Malenka RC, Quake SR, Südhof TC (2016) Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc Natl Acad Sci U S A 113:E5222–5231. 10.1073/pnas.1610155113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo MV, Földy C, Gokce O, Rothwell PE, Sun GL, Malenka RC, Südhof TC (2015) Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms. Neuron 87:326–340. 10.1016/j.neuron.2015.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K, Doty CD, Baek B, Ryu J, Sheng M (2013) Specific trans-synaptic interaction with inhibitory interneuronal neurexin underlies differential ability of neuroligins to induce functional inhibitory synapses. J Neurosci 33:3612–3623. 10.1523/JNEUROSCI.1811-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabernet L, Jadhav SP, Feldman DE, Carandini M, Scanziani M (2005) Somatosensory integration controlled by dynamic thalamocortical feed-forward inhibition. Neuron 48:315–327. 10.1016/j.neuron.2005.09.022 [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP; CommonMind Consortium, PsychENCODE Consortium, iPSYCH-BROAD Working Group (2018) Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359:693–697. 10.1126/science.aad6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LD, Cao Y, Pau G, Lawrence M, Wu TD, Seshagiri S, Gentleman R (2016) Prediction and quantification of splice events from RNA-seq data. PLoS One 11:e0156132. 10.1371/journal.pone.0156132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Cho RY, Lewis DA (2015) Alterations in cortical network oscillations and parvalbumin neurons in schizophrenia. Biol Psychiatry 77:1031–1040. 10.1016/j.biopsych.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Barry J, McDougel R, Terman D, Gu C (2012) Alternative splicing regulates kv3.1 polarized targeting to adjust maximal spiking frequency. J Biol Chem 287:1755–1769. 10.1074/jbc.M111.299305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackos DH, Lupardus PJ, Grand T, Chen Y, Wang TM, Reynen P, Gustafson A, Wallweber HJ, Volgraf M, Sellers BD, Schwarz JB, Paoletti P, Sheng M, Zhou Q, Hanson JE (2016) Positive allosteric modulators of GluN2A-containing NMDARs with distinct modes of action and impacts on circuit function. Neuron 89:983–999. 10.1016/j.neuron.2016.01.016 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S (2005) A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 3:e159. 10.1371/journal.pbio.0030159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, Sabatini BL, Greenberg ME (2018) Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21:120–129. 10.1038/s41593-017-0029-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T, Hansson L, Sønderby IE, Athanasiu L, Zuber V, Tesli M, Song J, Hultman CM, Bergen SE, Landén M, Melle I, Andreassen OA, Djurovic S (2016) A loss-of-function variant in a minor isoform of ANK3 protects against bipolar disorder and schizophrenia. Biol Psychiatry 80:323–330. 10.1016/j.biopsych.2015.09.021 [DOI] [PubMed] [Google Scholar]
- Hughes T, Sønderby IE, Polushina T, Hansson L, Holmgren A, Athanasiu L, Melbø-Jørgensen C, Hassani S, Hoeffding LK, Herms S, Bergen SE, Karlsson R, Song J, Rietschel M, Nöthen MM, Forstner AJ, Hoffmann P, Hultman CM, Landén M, Cichon S, et al. (2018) Elevated expression of a minor isoform of ANK3 is a risk factor for bipolar disorder. Transl Psychiatry 8:210. 10.1038/s41398-018-0175-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huusko N, Römer C, Ndode-Ekane XE, Lukasiuk K, Pitkänen A (2015) Loss of hippocampal interneurons and epileptogenesis: a comparison of two animal models of acquired epilepsy. Brain Struct Funct 220:153–191. 10.1007/s00429-013-0644-1 [DOI] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M (2011) How inhibition shapes cortical activity. Neuron 72:231–243. 10.1016/j.neuron.2011.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani MM, Jackson J, Ayzenshtat I, Hamzehei Sichani A, Manoocheri K, Kim S, Yuste R (2016) Opening holes in the blanket of inhibition: localized lateral disinhibition by VIP interneurons. J Neurosci 36:3471–3480. 10.1523/JNEUROSCI.3646-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepecs A, Fishell G (2014) Interneuron cell types are fit to function. Nature 505:318–326. 10.1038/nature12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshkhoo S, Vogt D, Sohal VS (2017) Dynamic, cell-type-specific roles for GABAergic interneurons in a mouse model of optogenetically inducible seizures. Neuron 93:291–298. 10.1016/j.neuron.2016.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B (2013) A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat Neurosci 16:1662–1670. 10.1038/nn.3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW (2012) Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci 35:57–67. 10.1016/j.tins.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich D, Winterer J, Que L, Luo W, Lukacsovich T, Földy C (2019) Single-cell RNA-seq reveals developmental origins and ontogenetic stability of neurexin alternative splicing profiles. Cell Rep 27:3752–3759.e4. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A, Centofante E, Bazinet JE, Tzeng CP, Mandel-Brehm C, Harmin DA, Adesnik H, Fagiolini M, Greenberg ME (2016) Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature 531:371–375. 10.1038/nature17187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O. (2012) Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci 13:107–120. 10.1038/nrn3155 [DOI] [PubMed] [Google Scholar]
- Meier J, Grantyn R (2004) Preferential accumulation of GABAA receptor gamma 2L, not gamma 2S, cytoplasmic loops at rat spinal cord inhibitory synapses. J Physiol 559:355–365. 10.1113/jphysiol.2004.066233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TM, Schreiner D, Xiao L, Traunmüller L, Bornmann C, Scheiffele P (2016) An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus. eLife 5:e22757. 10.7554/eLife.22757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Mucke L (2016) Network abnormalities and interneuron dysfunction in Alzheimer disease. Nat Rev Neurosci 17:777–792. 10.1038/nrn.2016.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ (2017) Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171:522–539.e20. 10.1016/j.cell.2017.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A (2013) Cortical interneurons that specialize in disinhibitory control. Nature 503:521–524. 10.1038/nature12676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Scanziani M (2001) Enforcement of temporal fidelity in pyramidal cells by somatic feed-forward inhibition. Science 293:1159–1163. 10.1126/science.1060342 [DOI] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott L, Andreassen OA, Cichon S, Craddock N, Edenberg HJ Jr, Nurnberger JI, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillen A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, et al. (2011) Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 43:977–983. 10.1038/ng.943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ (2015) Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron 87:14–27. 10.1016/j.neuron.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Ramos B, Baglietto-Vargas D, del Rio JC, Moreno-Gonzalez I, Santa-Maria C, Jimenez S, Caballero C, Lopez-Tellez JF, Khan ZU, Ruano D, Gutierrez A, Vitorica J (2006) Early neuropathology of somatostatin/NPY GABAergic cells in the hippocampus of a PS1xAPP transgenic model of Alzheimer's disease. Neurobiol Aging 27:1658–1672. 10.1016/j.neurobiolaging.2005.09.022 [DOI] [PubMed] [Google Scholar]
- Regan MC, Grant T, McDaniel MJ, Karakas E, Zhang J, Traynelis SF, Grigorieff N, Furukawa H (2018) Structural mechanism of functional modulation by gene splicing in NMDA receptors. Neuron 98:521–529.e3. 10.1016/j.neuron.2018.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J (2011) Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol 71:45–61. 10.1002/dneu.20853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert EH, Barker D, Ruderfer D, Bergen SE, O'Dushlaine C, Luce CJ, Sheridan SD, Theriault KM, Chambert K, Moran J, Purcell SM, Madison JM, Haggarty SJ, Sklar P (2013) Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Mol Psychiatry 18:922–929. 10.1038/mp.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Prybylowski K, Wang JF, Vicini S (2000) Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J Neurophysiol 83:1300–1306. 10.1152/jn.2000.83.3.1300 [DOI] [PubMed] [Google Scholar]
- Saxena A, Wagatsuma A, Noro Y, Kuji T, Asaka-Oba A, Watahiki A, Gurnot C, Fagiolini M, Hensch TK, Carninci P (2012) Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques 52:381–385. 10.2144/0000113878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study Consortium (2011) Genome-wide association study identifies five new schizophrenia loci. Nat Genet 43:969–976. 10.1038/ng.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid LC, Mittag M, Poll S, Steffen J, Wagner J, Geis HR, Schwarz I, Schmidt B, Schwarz MK, Remy S, Fuhrmann M (2016) Dysfunction of somatostatin-positive interneurons associated with memory deficits in an Alzheimer's disease model. Neuron 92:114–125. 10.1016/j.neuron.2016.08.034 [DOI] [PubMed] [Google Scholar]
- Schreiner D, Nguyen TM, Russo G, Heber S, Patrignani A, Ahrné E, Scheiffele P (2014) Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 84:386–398. 10.1016/j.neuron.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, Rossner MJ, Mann M, Simons M (2015) Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci 18:1819–1831. 10.1038/nn.4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Muñoz-Manchado AB, O'Donovan MC, Owen MJ, Pardiñas AF, Ryge J, Walters JTR, Linnarsson S, Lein ES; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Sullivan PF, Hjerling-Leffler J (2018) Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50:825–833. 10.1038/s41588-018-0129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K (2009) Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459:698–702. 10.1038/nature07991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH (1990) Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249:1580–1585. 10.1126/science.1699275 [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, Borneo J, Poon C, Ho T, Cai F, Steiner P, van der Brug MP, Modrusan Z, Kaminker JS, Hansen DV (2016) Untangling the brain's neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun 7:11295. 10.1038/ncomms11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455:903–911. 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai C, Abe Y, Westenbroek RE, Scheuer T, Catterall WA (2014) Impaired excitability of somatostatin- and parvalbumin-expressing cortical interneurons in a mouse model of Dravet syndrome. Proc Natl Acad Sci U S A 111:E3139–3148. 10.1073/pnas.1411131111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T. (2015) Interneuron dysfunction in syndromic autism: recent advances. Dev Neurosci 37:467–475. 10.1159/000434638 [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ (2011) A resource of cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71:995–1013. 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, Bertagnolli D, Goldy J, Shapovalova N, Parry S, Lee C, Smith K, Bernard A, Madisen L, Sunkin SM, Hawrylycz M, et al. (2016) Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19:335–346. 10.1038/nn.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A, Contestabile A, Ermolyuk YS, Ng T, Volynski KE, Soong TW, Goda Y, Cingolani LA (2017) Alternative splicing of P/Q-type Ca2+ channels shapes presynaptic plasticity. Cell Rep 20:333–343. 10.1016/j.celrep.2017.06.055 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF (1995) Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 268:873–876. 10.1126/science.7754371 [DOI] [PubMed] [Google Scholar]
- Treutlein B, Gokce O, Quake SR, Südhof TC (2014) Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci U S A 111:E1291–E1299. 10.1073/pnas.1403244111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban-Ciecko J, Barth AL (2016) Somatostatin-expressing neurons in cortical networks. Nat Rev Neurosci 17:401–409. 10.1038/nrn.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret L, Mann EO, Hang GB, Barth AM, Cobos I, Ho K, Devidze N, Masliah E, Kreitzer AC, Mody I, Mucke L, Palop JJ (2012) Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in Alzheimer model. Cell 149:708–721. 10.1016/j.cell.2012.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veys K, Snyders D, De Schutter E (2013) Kv3.3b expression defines the shape of the complex spike in the Purkinje cell. Front Cell Neurosci 7:205. 10.3389/fncel.2013.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamsley B, Jaglin XH, Favuzzi E, Quattrocolo G, Nigro MJ, Yusuf N, Khodadadi-Jamayran A, Rudy B, Fishell G (2018) Rbfox1 mediates cell-type-specific splicing in cortical interneurons. Neuron 100:846–859.e7. 10.1016/j.neuron.2018.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting P, McKernan RM, Iversen LL (1990) Another mechanism for creating diversity in gamma-aminobutyrate type A receptors: RNA splicing directs expression of two forms of gamma 2 phosphorylation site. Proc Natl Acad Sci U S A 87:9966–9970. 10.1073/pnas.87.24.9966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, Hong W (2017) Detecting activated cell populations using single-cell RNA-seq. Neuron 96:313–329.e6. 10.1016/j.neuron.2017.09.026 [DOI] [PubMed] [Google Scholar]
- Xu X, Wells AB, O'Brien DR, Nehorai A, Dougherty JD (2014) Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci 34:1420–1431. 10.1523/JNEUROSCI.4488-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S (2015) Brain structure. cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347:1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-seq quality metrics RNA-sequencing quality control metrics across the 24 samples generated in this study (a). Samples are in columns. QC metrics are shown in rows. Rows contain different quality control metrics used to assess each sample. Descriptions of quality control metrics are indicated by text labeling each row. 5’/3’ coverage plots for the current study (b) and the samples from GSE77243 (c) and GSE71585 (d) included for comparison in our website resource. Briefly, for each gene g a coverage vector cgi was calculated giving the number of reads overlapping position i of the gene, for i=1...ng tracking the annotated exonic bases of the gene from 1 to the total number of exonic bases ng of gene g, from 5’ (i = 1) to 3’ (i=ng) in the direction of transcription. Then the gene was divided into 20 equally spaced bins Bb of ng/20 bases each. The average coverage acgb was then calculated as the average over all the bases in the bin of cgi, or acgb=1/(ng/20) ∑iEBb cgi . Next, these average coverages acgb were converted into normalized coverages ncgb by dividing by the average coverage over the entire gene: acg=1/ng ∑(i=1)(ng)cgi and then ncgb=acgb/acg . This normalized coverage ncgb is then a matrix whose rows are all the genes and columns are the twenty bins. For speed of calculation and robustness, we selected 500 genes at random among those that were expressed with at least 4 nRPKMs in at least 1/4 of the samples. Finally, we calculated the median normalized coverage for each bin over those 500 genes: ncb=“median”gE“500 genes” (ncgb). This statistic is plotted as the Median Relative Coverage. Download Figure 1-1, PDF file (624KB, pdf)
Differential gene expression Differential expression results per gene, including average RPKM expression levels per neuron subtype per gene, and neuron subtype pairwise log2FC and p-value statistics. Download Figure 1-2, PDF file (624KB, pdf)
Druggable targets in single neuron subtypes Druggable targets enriched in each neuron subtype, including druggable genome annotations. Download Figure 1-3, PDF file (624KB, pdf)
Druggable targets in pairs of neuron subtypes Druggable targets enriched in pairs of neuron subtypes, including druggable genome annotations. Download Figure 1-4, PDF file (624KB, pdf)
Differentially spliced events in single neuron subtypes Differentially spliced events in a single neuron subtype and corresponding log(fold change) and p-value statistics. Variant types are defined as follows: SE (skipped exon), S2E (two consecutive exons skipped), RI (retained intron), MXE (mutually exclusive exons), A5SS (alternative 5’ splice site), A3SS (alternative 3’ splice site), AFE (alternative first exon), ALE (alternative last exon), AS (alternative start other than AFE), AE (alternative end other than ALE). Download Figure 2-1, PDF file (1.9MB, pdf)
Differentially spliced events in pairs of neuron subtypes Differentially spliced events in two neuron subtypes and corresponding log(fold change) and p-value statistics. Variant types are defined as in Figure 2-1. Download Figure 2-2, PDF file (1.9MB, pdf)
Example sequencing coverage at splice junctions IGV genome browser screenshots of the sequencing coverage at and surrounding the splice junctions featured in Figure 4. In the genome browser screenshots, each row represents the read coverage of a single sample. The first 6 red samples are the EXC samples, followed by 6 green PV samples, then 6 purple SST samples, and 6 orange VIP samples. Read counts of splice junctions corresponding to the four events featured in Figure 3 (e-h). In the splice read count plots, each row shows counts of reads by sample for a specific neuronal population isolated. Read counts reflect the total count of reads supporting and spanning a splice event variant, as indicated in methods. Gene models were then mapped to the same genomic coordinates for display in the figure. Each sample is represented with a different colored line as indicated in the key to the right of each row. In the bottom panel, exons are indicated by boxes and introns by lines. Download Figure 4-1, XLS file (15.2MB, xls)