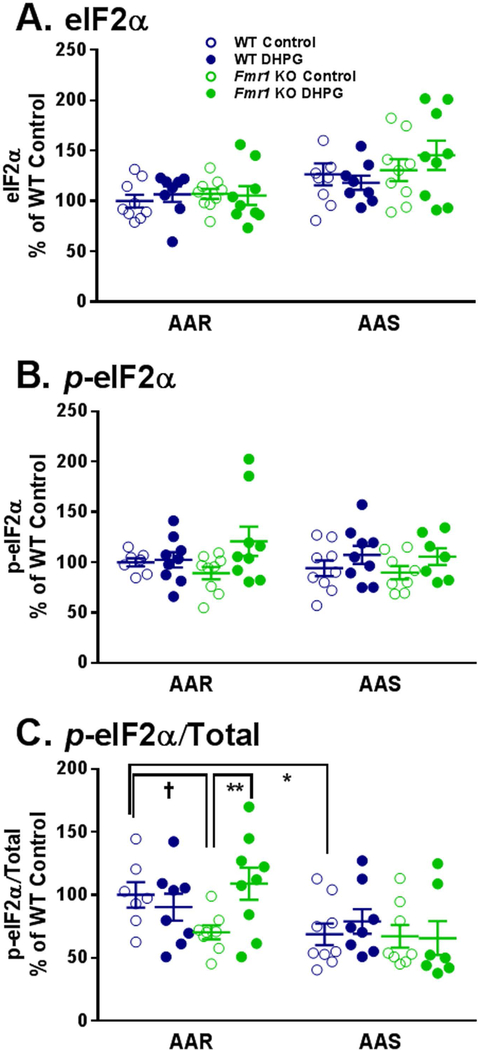
Fig. 4.
Effect of treatment with DHPG (100 μM) on levels of eIF2α (A), p-eIF2α (B), and p-eIF2α/Total (C) in hippocampal slice preparations from WT and Fmr1 KO mice in AAR and AAS conditions. Measurements were made in nine mice for each group. A. One WT DHPG treated slice was excluded from both the AAR and the AAS conditions as an outlier. B. In the AAR condition, two WT Control slices and in the AAS condition one Fmr1 KO DHPG treated slice were excluded due to artifacts on Western blots. In AAS, one Fmr1 KO Control slice and one Fmr1 KO DHPG treated slice were excluded as outliers. C. All exclusions were a result of prior exclusions of either the p-eIF2α or total eIF2α. In the AAR condition, two WT Control slices and one WT DHPG treated slice were excluded. In the AAS condition, one WT DHPG treated slice, one Fmr1 KO Control slice, and two Fmr1 KO DHPG treated slices were excluded. Data were analyzed by means of a three-way ANOVA with amino acid condition (AAR, AAS), genotype (WT, Fmr1 KO), and treatment (+/− DHPG) as between subject factors (Table 1). A. For eIF2α, the main effect of amino acid condition was statistically significant (p < 0.001), indicating that regardless of genotype or treatment levels of eIF2α are higher in AAS conditions. B. For p-eIF2α, the main effect of treatment was statistically significant (p = 0.014), indicating that regardless of genotype or amino acid conditions DHPG resulted in an increase in p-eIF2α. C. For p-eIF2α/Total, the amino acid condition x genotype x treatment interaction was statistically significant (p = 0.044). We probed for individual group and condition differences by means of Bonferroni-corrected post hoc t-tests. Statistically significant effects are indicated on the figure: †, 0.05 ≤ p ≤ 0.10; *, 0.01 ≤ p ≤ 0.05; **, 0.001 ≤ p ≤ 0.01.