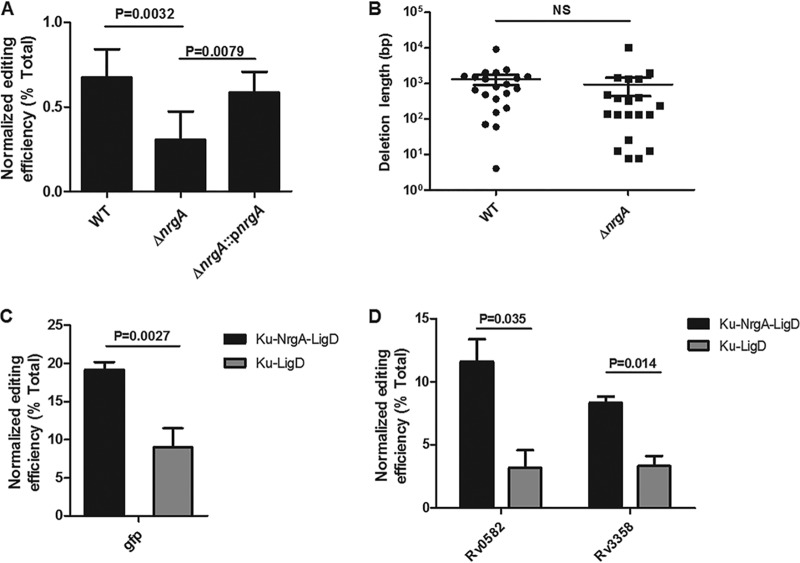
FIG 3.
NrgA is involved in CRISPR-NHEJ genome editing in mycobacteria. (A) Genome-editing efficiency in M. marinum derivatives. The whiB6 crRNA plasmid (pYC1103) was transformed into the wild-type, nrgA mutant, or nrgA-complemented strain, and 24 colonies in each group were picked for PCR and sequencing analysis. Editing efficiency was calculated as the ratio of the number of edited events to the total number of colonies tested by PCR. Normalized editing efficiency was calculated as the editing efficiency × (total CFU obtained with whiB6 targeting sgRNA/total CFU obtained with control sgRNA). (B) Deletion length distributions of whiB6 gene in wild-type and nrgA mutant strains in one experiment. Bars represent median deletion size for each strain. NS, not significant. (C, D) NrgA increases the CRISPR-NHEJ genome-editing efficiency in M. smegmatis (C) and M. tuberculosis (D). crRNA plasmids were electroporated into M. smegmatis and M. tuberculosis cells with plasmids expressing the complete NHEJ machinery (pNHEJ-Cas12a-recX for M. smegmatis and pNHEJ-recX for M. tuberculosis; Ku-NrgA-LigD) or the NHEJ machinery without NrgA (pYC1376 for M. smegmatis and pYC1654 for M. tuberculosis; Ku-LigD). For M. smegmatis, a chromosomally integrated gfp reporter gene was edited. Normalized efficiency was calculated as the frequency of GFP-negative (white) transformants × (total CFU obtained with gfp-targeting sgRNA/total CFU obtained with control sgRNA). For M. tuberculosis, 24 colonies in each group were picked for PCR and sequencing analysis. Normalized editing efficiency was calculated as the editing efficiency × (total CFU obtained with target sgRNA/total CFU obtained with control sgRNA). (A, C, D) Bars represent mean values ± standard deviations from three independent experiments. P values were determined via Student’s unpaired t test.